ATP binding domain 4 Introductions: Difference between revisions
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
It was found that ATP pyrophosphatase involved in hydrolyzing the phosphodiester bond in ATP. We have been assigned to investigate ATP Binding Domain 4 (1ru8A) which is a N-type ATP pyrophosphatase from Pyrococcus furiosus. N-type ATP pyrophosphatase is a family belongs to the superfamily- Adenine Nucleotide Alpha Hydrolases-like (AANH-like). Other members of AANH-like are arginosuccinate synthase, NAD synthase, GMP synthase and tRNA Methyl transferase. All these family members contain highly conserved PP-loop which is likely to be involved in phosphate binding. This motif is a modified form of P-loop of nucleotide binding domains. PP-loop consists of conserved motif SGGKD at the N terminus. These sequences are highly conserved among all the family members (See '''Diagram 2''') and across all species in domain Archaea and Eukarya but not bacteria. Based on the comparison between other family members such as arginosuccinate synthase, the function of ATP binding Domain 4 can be inferred. The function can be inferred based on the similarity of the structure and evolution of the amino acid sequences. | It was found that ATP pyrophosphatase involved in hydrolyzing the phosphodiester bond in ATP. We have been assigned to investigate ATP Binding Domain 4 (1ru8A) which is a N-type ATP pyrophosphatase from ''Pyrococcus furiosus''. N-type ATP pyrophosphatase is a family belongs to the superfamily- Adenine Nucleotide Alpha Hydrolases-like (AANH-like). Other members of AANH-like are arginosuccinate synthase, NAD synthase, GMP synthase and tRNA Methyl transferase. All these family members contain highly conserved PP-loop which is likely to be involved in phosphate binding. This motif is a modified form of P-loop of nucleotide binding domains. PP-loop consists of conserved motif SGGKD at the N terminus. These sequences are highly conserved among all the family members (See '''Diagram 2''') and across all species in domain Archaea and Eukarya but not bacteria. Based on the comparison between other family members such as arginosuccinate synthase, the function of ATP binding Domain 4 can be inferred. The function can be inferred based on the similarity of the structure and evolution of the amino acid sequences. | ||
Revision as of 05:06, 2 June 2009
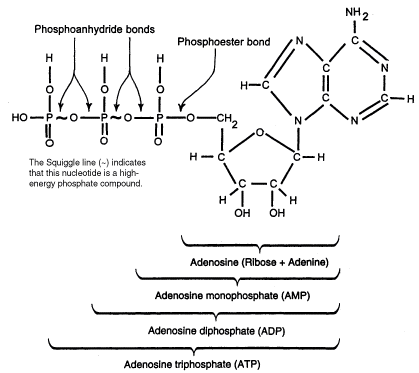
ATP consists of adenosine nucleotide linked to 3 phosphate groups. The third phosphate group of ATP can be removed by hydrolysis thus releasing free energy making it a major energy currency of the cell. ATP can be consumed by cells which undergo anabolic reaction such as cell division, active transport and muscle contraction.
Diagram 1.ATP cleavage.
It was found that ATP pyrophosphatase involved in hydrolyzing the phosphodiester bond in ATP. We have been assigned to investigate ATP Binding Domain 4 (1ru8A) which is a N-type ATP pyrophosphatase from Pyrococcus furiosus. N-type ATP pyrophosphatase is a family belongs to the superfamily- Adenine Nucleotide Alpha Hydrolases-like (AANH-like). Other members of AANH-like are arginosuccinate synthase, NAD synthase, GMP synthase and tRNA Methyl transferase. All these family members contain highly conserved PP-loop which is likely to be involved in phosphate binding. This motif is a modified form of P-loop of nucleotide binding domains. PP-loop consists of conserved motif SGGKD at the N terminus. These sequences are highly conserved among all the family members (See Diagram 2) and across all species in domain Archaea and Eukarya but not bacteria. Based on the comparison between other family members such as arginosuccinate synthase, the function of ATP binding Domain 4 can be inferred. The function can be inferred based on the similarity of the structure and evolution of the amino acid sequences.
Diagram 2. Summary of enzymes containing the PP-loop motif. Adapted from Bork. P.& Koonin, E.V.
Back to Main ATP binding domain 4 pages
Bork, P & Koonin, E.V. (1994), A P-Loop-Like Motif in a Widespread ATP Pyrophosphatase Domain: Implications for the Evolution of Sequence Motifs and Enzyme Activity, Proteins: Structure, Function and Genetics, 20, 347-355.