Structure of N-acetylneuraminic acid phosphatase: Difference between revisions
JasonCheong (talk | contribs) No edit summary |
|||
| (60 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__TOC__ | __TOC__ | ||
=== | ===Chain Sequence=== | ||
MGSDKIHHHHHHMGLSRVRAVFFDLDNTLIDTAGASRRGMLEVIKLLQSKYHYKEEAEIICDKVQVKLSKECFHPYSTCI | MGSDKIHHHHHHMGLSRVRAVFFDLDNTLIDTAGASRRGMLEVIKLLQSKYHYKEEAEIICDKVQVKLSKECFHPYSTCI | ||
TDVRTSHWEEAIQETKGGADNRKLAEECYFLWKSTRLQHMILADDVKAMLTELRKEVRLLLLTNGDRQTQREKIEACACQ | TDVRTSHWEEAIQETKGGADNRKLAEECYFLWKSTRLQHMILADDVKAMLTELRKEVRLLLLTNGDRQTQREKIEACACQ | ||
| Line 8: | Line 8: | ||
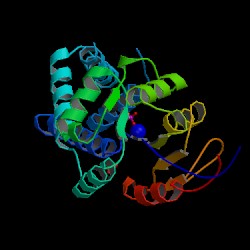
[[Image:2gfh asym r 250.jpg|thumb|structure 18|left]] | [[Image:2gfh asym r 250.jpg|thumb|structure 18|left]] | ||
[[Image:Interpro1.JPG|interpro|left]] | |||
| Line 59: | Line 89: | ||
===DALI | ===DALI Search Results=== | ||
SUMMARY: PDB/chain identifiers and structural alignment statistics | SUMMARY: PDB/chain identifiers and structural alignment statistics | ||
| Line 98: | Line 126: | ||
29: 3033-A 1mh9-A 9.2 3.2 146 194 15 0 0 15 S HYDROLASE deoxyribonucleotidase (mitochondrial 5'(3')- | 29: 3033-A 1mh9-A 9.2 3.2 146 194 15 0 0 15 S HYDROLASE deoxyribonucleotidase (mitochondrial 5'(3')- | ||
=== | ===Surface properties=== | ||
[[Image:Surface properties.JPG]] | |||
===From PDB=== | |||
Chemical component identifier would be | |||
PO4 PHOSPHATE ION | |||
NA SODIUM ION | |||
EDO 1,2-ETHANEDIOL | |||
CL CHLORIDE ION | |||
Closely related to 2HO4,PO4 PHOSPHATE ION found. | |||
Cloesly related to 2HX1,CL Chloride Ion, EDO 1 2 Ethanediol found. | |||
'''2GFH''' | |||
http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=2GFH | |||
'''2HX1''' | |||
http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=2HX1 | |||
'''1FEZ''' | |||
http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=1FEZ | |||
'''2HO4''' | |||
http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=2HO4 | |||
===Pubmed=== | |||
The identity of Asp12 as the active-site nucleophile was further evidenced by the observed removal of catalytic activity resulting from Asp12Ala substitution | |||
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=10956028 | |||
===Pfarm classification=== | |||
PFAM Accession:PF00702 | |||
PFAM ID:hydrolase | |||
http://www.sanger.ac.uk//cgi-bin/Pfam/getacc?PF00702 | |||
===CE=== | |||
'''2GFH''' USR1:A(size=260) vs '''1FEZ''' USR2:A(size=256) | |||
Structure Alignment | |||
'''2GFH''' USR1:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 4; CHAIN: A; ENGINEERED: YES | |||
'''1FEZ''' USR2:A - MOL_ID: 1; MOLECULE: PHOSPHONOACETALDEHYDE HYDROLASE; CHAIN: A, B, C, D; ENGINEERED: YES | |||
USR1:A 17/6 RVRAVFFDLDNTLID-TAGASRRGMLEVIKLLQSKYHYKEEAEIICDKVQVKLSKECFHP | |||
USR2:A 1/6 KIEAVIFDWAGTTVDYGCFAPLEVFMEIFHKRG--------VAITAEEARKPMGLL---- | |||
USR1:A 76/#12 YSTCITDVRTSHWEEA--IQETKG---GADN--RKLAEECYFLWKST--RLQHMILADDV | |||
USR2:A 49/54 -----KIDHVRALTEMPRIASEWNRVFRQLPTEADIQEMYEEFEEILFAILPRYASPINA | |||
USR1:A 127/116 KAMLTELRK--EVRLLLLTNGDRQTQREKIEACACQ-----SYFDAIVIGGEQKEEKPAP | |||
USR2:A 104/109 VKEVIASLRERGIKIGSTTG----YTREMMDIVAKEAALQGYKPDFLVTPDDVPAGRPYP | |||
USR1:A 180/169 SIFYHCCDLLGVQP-GDCVMVGDTLETDIQGGLNAGLKATVWINK--------------- | |||
USR2:A 160/165 WMCYKNAMELGVYPMNHMIKVGDTVSDMKEGRNAGM--WTVGVILGSSELGLTEEEVENM | |||
USR1:A 224/213 --------SGR-VPLTSSPMPHYMVSSVLELPALLQSI | |||
USR2:A 218/223 DSVELREKIEVVRNRFVENGAHFTIETMQELESVMEHI | |||
'''2gfh''' USR1:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 4; CHAIN: A; ENGINEERED: YES | |||
'''2hx1''' USR2:A - MOL_ID: 1; MOLECULE: PREDICTED SUGAR PHOSPHATASES OF THE HAD SUPERFAMILY; CHAIN: A, B, C, D; ENGINEERED: YES | |||
USR1:A 65/54 QVKLSKECFHPYST------------------------------CITDVRTSHWEEAIQE | |||
USR2:A 24/24 VLKTYNGL------LPGIENTFDYLKAQGQDYYIVTNDASRSPEQLADSYHKL------- | |||
USR1:A 95/84 TKGGADNRKLAEECYFLWKSTRLQHM---ILADDVKAMLTELRKEVRLLLLTNGDRQTQR | |||
USR2:A 71/71 ----------GLFSI-TADKIISSGXITKEYIDLKVDGG-------IVAYLG------TA | |||
USR1:A 152/141 EKIEACACQSYFDAIV-----------------IGGEQKEEKPAPSIFYHCCDLLGVQPG | |||
USR2:A 107/107 NSANY-LVSDGIKXLPVSAIDDSNIGEVNALVLLDDEGF------NWFHDLNKTVNLLRK | |||
USR1:A 195/184 DC-VMVGDTL---------------ETDIQGGLNAGLKATVWI | |||
USR2:A 160/160 RTIPAIVANTDNTYPLTKTDVAIAIGGVATXIES-ILGRRFIR | |||
'''2gfh''' USR1:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 4; CHAIN: A; ENGINEERED: YES | |||
'''2ho4''' USR2:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 2; CHAIN: A, B; ENGINEERED: YES | |||
Model | USR1:A 122/111 LADDVKAMLTELRKEV--RLLLLTNGDRQTQREKIEACACQSYFD---------AIVIGG | ||
USR2:A 71/72 EIFTSLTAARNLIEQKQVRPXLLLD-----------DRALPEFTGVQTQDPNAVVIGLAP | |||
USR1:A 171/160 EQKEEKPAPSIFYHCCDLLGVQPGDCVMVGD-------------TLETDIQGGLNAGLKA | |||
USR2:A 120/121 EH----FHYQLLNQAFRLLLDG--APLIAIHKARYYKRKDGLALGPGPFVTALEYATDTK | |||
USR1:A 218/207 TVWINKSGRVPLTSSPMPHYMVSSVLELP | |||
USR2:A 174/175 AXV--------------------VGKPEK | |||
[[Image:2gfh and 2hx1.JPG]] | |||
2gfh vs 2hx1 Rmsd = 5.6Å Z-Score = 3.1 | |||
Sequence identity = 6.2% | |||
Aligned/gap positions = 112/111 | |||
[[Image:2gfh and 1fez.JPG]] | |||
2gfh vs 1fez Rmsd = 4.4Å Z-Score = 5.5 | |||
Sequence identity = 17.4% | |||
Aligned/gap positions = 213/65 | |||
[[Image:2gfh and 2ho4.JPG]] | |||
2gfh vs 2ho4 Rmsd = 5.0Å Z-Score = 3.3 | |||
Sequence identity = 11.4% | |||
Aligned/gap positions = 88/61 | |||
===ProFunc=== | |||
http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/profunc/GetResults.pl?source=profunc&user_id=bb32&code=143144 | |||
[[Image:Conserved.JPG]] | |||
Diagram showing the conserved region ASN15 , Thr16, Leu17 with EDO4 molecule in grey and PO4 in yellow. | |||
[[Image:Po4 ligand.JPG]] | |||
IMPORTANT!!! This is involved in the function. Most likely a phosphatase. | |||
[[Image:Edo4 ligand.JPG]] | |||
Most likely an antifreezer. | |||
[[Image:Clefts.JPG]] | |||
Possible binding sites. Results shows 2 ligands PO4 and EDO4 | |||
===Pfam 21.0 (Janelia Farm)Alignments of top-scoring domains=== | |||
{| border="1" cellpadding="15" cellspacing="0" | |||
|+'''Table of functions''' | |||
|Model | |||
Hydrolase | |||
| | | | ||
Seq-from:18 | Seq-from:18 | ||
| Line 138: | Line 300: | ||
+ +++ + +AG+++++++n + | + +++ + +AG+++++++n + | ||
query 203 LETDIQGGLnAGLkATVWINKS 224 | query 203 LETDIQGGLnAGLkATVWINKS 224 | ||
|- | |||
|} | |||
===External Link=== | |||
[http://www.ebi.ac.uk/msd-srv/msdlite/atlas/summary/2gfh.html 1. EBI Mcromolecular Structure Database] | |||
[http://www.ebi.ac.uk/cgi-bin/iprscan/iprscan?tool=iprscan&jobid=iprscan-20070515-06405113 2. InterProScan] | |||
---- | |||
Return to '''[http://compbio.chemistry.uq.edu.au/mediawiki/index.php/N-acetylneuraminic_acid_phosphatase Home Page]''' | |||
Latest revision as of 01:26, 12 June 2007
Chain Sequence
MGSDKIHHHHHHMGLSRVRAVFFDLDNTLIDTAGASRRGMLEVIKLLQSKYHYKEEAEIICDKVQVKLSKECFHPYSTCI TDVRTSHWEEAIQETKGGADNRKLAEECYFLWKSTRLQHMILADDVKAMLTELRKEVRLLLLTNGDRQTQREKIEACACQ SYFDAIVIGGEQKEEKPAPSIFYHCCDLLGVQPGDCVMVGDTLETDIQGGLNAGLKATVWINKSGRVPLTSSPMPHYMVS SVLELPALLQSIDCKVSMSV
DALI Search Results
SUMMARY: PDB/chain identifiers and structural alignment statistics NR. STRID1 STRID2 Z RMSD LALI LSEQ2 %IDE REVERS PERMUT NFRAG TOPO PROTEIN 1: 3033-A 2gfh-A 41.1 0.0 246 246 100 0 0 1 S HYDROLASE haloacid dehalogenase-like hydrolase domain 2: 3033-A 1fez-A 18.1 3.5 178 256 22 0 0 13 S HYDROLASE phosphonoacetaldehyde hydrolase (bacillus c 3: 3033-A 2hsz-A 17.9 3.3 168 222 23 0 0 13 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION novel predicted 4: 3033-A 1qq5-A 17.3 3.1 198 245 19 0 0 12 S HYDROLASE l-2-haloacid dehalogenase (xanthobacter aut 5: 3033-A 1o03-A 17.0 5.0 188 221 20 0 0 11 S ISOMERASE beta-phosphoglucomutase (lactococcus lactis 6: 3033-A 2b0c-A 16.4 2.6 184 199 20 0 0 13 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION putative phospha 7: 3033-A 2fdr-A 15.8 4.4 190 214 19 0 0 15 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION conserved hypoth 8: 3033-A 2p11-A 15.7 2.9 194 211 16 0 0 20 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION hypothetical pro 9: 3033-A 1te2-A 15.7 3.6 170 211 19 0 0 15 S HYDROLASE putative phosphatase (escherichia coli o157 10: 3033-A 1yns-A 15.3 4.0 169 254 11 0 0 13 S HYDROLASE e-1 enzyme (enolase-phosphatase e1) (homo s 11: 3033-A 1qyi-A 15.0 3.5 198 375 19 0 0 17 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION hypothetical pro 12: 3033-A 2i6x-A 14.9 3.1 176 199 19 0 0 18 S HYDROLASE hydrolase, haloacid dehalogenase-like family 13: 3033-A 1u7p-A 14.3 2.9 144 164 18 0 0 14 S HYDROLASE magnesium-dependent phosphatase-1 (mdp-1) ( 14: 3033-A 1ymq-A 14.1 2.3 130 260 16 0 0 14 S TRANSFERASE sugar-phosphate phosphatase bt4131 (bacte 15: 3033-A 1j8d-A 13.1 2.5 141 180 11 0 0 12 S HYDROLASE deoxy-d-mannose-octulosonate 8-phosphate ph 16: 3033-A 2ho4-A 12.9 2.4 131 246 19 0 0 14 S HYDROLASE haloacid dehalogenase-like hydrolase domain 17: 3033-A 1pw5-A 12.7 2.3 136 246 21 0 0 12 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION nagd protein, pu 18: 3033-A 1nf2-A 12.7 2.6 127 267 13 0 0 11 S STRUCTURAL GENOMICS/UNKNOWN FUNCTION phosphatase (the 19: 3033-A 1rlm-A 12.4 2.8 131 269 13 0 0 14 S HYDROLASE phosphatase Mutant (escherichia coli) bacte 20: 3033-A 1f5s-A 12.1 3.5 159 210 14 0 0 15 S HYDROLASE phosphoserine phosphatase (psp) (methanoco 21: 3033-A 1cr6-B 12.0 3.8 177 541 18 0 0 18 S HYDROLASE epoxide hydrolase (mus musculus) mouse expr 22: 3033-A 1rku-A 11.9 3.6 172 206 11 0 0 18 S TRANSFERASE homoserine kinase (pseudomonas aeruginosa 23: 3033-A 2b30-A 11.8 2.7 134 284 16 0 0 12 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION pvivax hypotheti 24: 3033-A 1kyt-A 10.5 2.5 122 216 13 0 0 15 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION hypothetical pro 25: 3033-A 2o2x-A 10.3 3.6 139 204 17 0 0 14 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION hypothetical pro 26: 3033-A 1u02-A 10.1 2.7 128 222 16 0 0 12 S STRUCTURAL GENOMICS trehalose-6-phosphate phosphatase 27: 3033-A 2fea-A 10.0 3.5 167 219 7 0 0 21 S HYDROLASE 2-hydroxy-3-keto-5-methylthiopentenyl-1- pho 28: 3033-A 2hx1-A 9.6 3.2 130 275 24 0 0 19 S HYDROLASE predicted sugar phosphatases of the had supe 29: 3033-A 1mh9-A 9.2 3.2 146 194 15 0 0 15 S HYDROLASE deoxyribonucleotidase (mitochondrial 5'(3')-
Surface properties
From PDB
Chemical component identifier would be
PO4 PHOSPHATE ION
NA SODIUM ION
EDO 1,2-ETHANEDIOL
CL CHLORIDE ION
Closely related to 2HO4,PO4 PHOSPHATE ION found.
Cloesly related to 2HX1,CL Chloride Ion, EDO 1 2 Ethanediol found.
2GFH http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=2GFH
2HX1 http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=2HX1
1FEZ http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=1FEZ
2HO4 http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=2HO4
Pubmed
The identity of Asp12 as the active-site nucleophile was further evidenced by the observed removal of catalytic activity resulting from Asp12Ala substitution http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Abstract&list_uids=10956028
Pfarm classification
PFAM Accession:PF00702
PFAM ID:hydrolase
http://www.sanger.ac.uk//cgi-bin/Pfam/getacc?PF00702
CE
2GFH USR1:A(size=260) vs 1FEZ USR2:A(size=256) Structure Alignment
2GFH USR1:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 4; CHAIN: A; ENGINEERED: YES
1FEZ USR2:A - MOL_ID: 1; MOLECULE: PHOSPHONOACETALDEHYDE HYDROLASE; CHAIN: A, B, C, D; ENGINEERED: YES
USR1:A 17/6 RVRAVFFDLDNTLID-TAGASRRGMLEVIKLLQSKYHYKEEAEIICDKVQVKLSKECFHP USR2:A 1/6 KIEAVIFDWAGTTVDYGCFAPLEVFMEIFHKRG--------VAITAEEARKPMGLL---- USR1:A 76/#12 YSTCITDVRTSHWEEA--IQETKG---GADN--RKLAEECYFLWKST--RLQHMILADDV USR2:A 49/54 -----KIDHVRALTEMPRIASEWNRVFRQLPTEADIQEMYEEFEEILFAILPRYASPINA USR1:A 127/116 KAMLTELRK--EVRLLLLTNGDRQTQREKIEACACQ-----SYFDAIVIGGEQKEEKPAP USR2:A 104/109 VKEVIASLRERGIKIGSTTG----YTREMMDIVAKEAALQGYKPDFLVTPDDVPAGRPYP USR1:A 180/169 SIFYHCCDLLGVQP-GDCVMVGDTLETDIQGGLNAGLKATVWINK--------------- USR2:A 160/165 WMCYKNAMELGVYPMNHMIKVGDTVSDMKEGRNAGM--WTVGVILGSSELGLTEEEVENM USR1:A 224/213 --------SGR-VPLTSSPMPHYMVSSVLELPALLQSI USR2:A 218/223 DSVELREKIEVVRNRFVENGAHFTIETMQELESVMEHI
2gfh USR1:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 4; CHAIN: A; ENGINEERED: YES
2hx1 USR2:A - MOL_ID: 1; MOLECULE: PREDICTED SUGAR PHOSPHATASES OF THE HAD SUPERFAMILY; CHAIN: A, B, C, D; ENGINEERED: YES
USR1:A 65/54 QVKLSKECFHPYST------------------------------CITDVRTSHWEEAIQE USR2:A 24/24 VLKTYNGL------LPGIENTFDYLKAQGQDYYIVTNDASRSPEQLADSYHKL------- USR1:A 95/84 TKGGADNRKLAEECYFLWKSTRLQHM---ILADDVKAMLTELRKEVRLLLLTNGDRQTQR USR2:A 71/71 ----------GLFSI-TADKIISSGXITKEYIDLKVDGG-------IVAYLG------TA USR1:A 152/141 EKIEACACQSYFDAIV-----------------IGGEQKEEKPAPSIFYHCCDLLGVQPG USR2:A 107/107 NSANY-LVSDGIKXLPVSAIDDSNIGEVNALVLLDDEGF------NWFHDLNKTVNLLRK USR1:A 195/184 DC-VMVGDTL---------------ETDIQGGLNAGLKATVWI USR2:A 160/160 RTIPAIVANTDNTYPLTKTDVAIAIGGVATXIES-ILGRRFIR
2gfh USR1:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 4; CHAIN: A; ENGINEERED: YES
2ho4 USR2:A - MOL_ID: 1; MOLECULE: HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN CONTAINING 2; CHAIN: A, B; ENGINEERED: YES
USR1:A 122/111 LADDVKAMLTELRKEV--RLLLLTNGDRQTQREKIEACACQSYFD---------AIVIGG USR2:A 71/72 EIFTSLTAARNLIEQKQVRPXLLLD-----------DRALPEFTGVQTQDPNAVVIGLAP USR1:A 171/160 EQKEEKPAPSIFYHCCDLLGVQPGDCVMVGD-------------TLETDIQGGLNAGLKA USR2:A 120/121 EH----FHYQLLNQAFRLLLDG--APLIAIHKARYYKRKDGLALGPGPFVTALEYATDTK USR1:A 218/207 TVWINKSGRVPLTSSPMPHYMVSSVLELP USR2:A 174/175 AXV--------------------VGKPEK
2gfh vs 2hx1 Rmsd = 5.6Å Z-Score = 3.1 Sequence identity = 6.2% Aligned/gap positions = 112/111
2gfh vs 1fez Rmsd = 4.4Å Z-Score = 5.5 Sequence identity = 17.4% Aligned/gap positions = 213/65
2gfh vs 2ho4 Rmsd = 5.0Å Z-Score = 3.3 Sequence identity = 11.4% Aligned/gap positions = 88/61
ProFunc
Diagram showing the conserved region ASN15 , Thr16, Leu17 with EDO4 molecule in grey and PO4 in yellow.
IMPORTANT!!! This is involved in the function. Most likely a phosphatase.
Most likely an antifreezer.
Possible binding sites. Results shows 2 ligands PO4 and EDO4
Pfam 21.0 (Janelia Farm)Alignments of top-scoring domains
| Model
Hydrolase |
Seq-from:18 |
Seq-to:224 |
HMM-from:1 |
HMM-to:183 |
Score:96.2 |
E-value:1e-25 |
Alignment:glocal |
Description:haloacid dehalogenase-like hydrolase Hydrolase: domain 1 of 1, from 18 to 224: score 96.2, E = 1e-25
*->ikavvFDkDGTLtdgkeppiaeaiveaaaelgl.........lplee
++av+FD+D+TL+d+ + + ++ + e+ ++l + + +++ ++ +
query 18 VRAVFFDLDNTLIDT-AGASRRGMLEVIKLLQSkyhykeeaeIICDK 63
vekllgrgl.g.erilleggltaell...................d.evl
v l +++ ++ ++ t ++ + +++++ ++++ ++ ++
query 64 VQVKLSKECfHpYSTCITDVRTSHWEeaiqetkggadnrklaeecYfLWK 113
glial.dklypgarealkaLkrrGikvailTggdr.naeallealgla.l
++ ++ l +++++ l +L++ +++ +lT+gdr++++++ ea+++ ++
query 114 STRLQhMILADDVKAMLTELRKE-VRLLLLTNGDRqTQREKIEACACQsY 162
fdviidsdevggvgpivvgKPkpeifllalerlgvkpeevgpevlmVGDg
fd+i++++e + KP+p if + ++ lgv+p ++ +mVGD+
query 163 FDAIVIGGEQK------EEKPAPSIFYHCCDLLGVQPGDC----VMVGDT 202
vnDapalaa.AGv.gvamgngg<-*
+ +++ + +AG+++++++n +
query 203 LETDIQGGLnAGLkATVWINKS 224
|
External Link
1. EBI Mcromolecular Structure Database
Return to Home Page