|
|
| (32 intermediate revisions by 3 users not shown) |
| Line 1: |
Line 1: |
| {{User:Cacycle/wikEd userbox}}
| | The predicted function based on the evolution and structure |
|
| |
|
| | <font size = "4">'''Hydrolase'''</font> |
|
| |
|
| __TOC__
| |
|
| |
|
| =='''Proposed Functions'''==
| | [[Image:Document18_01.png]] |
|
| |
|
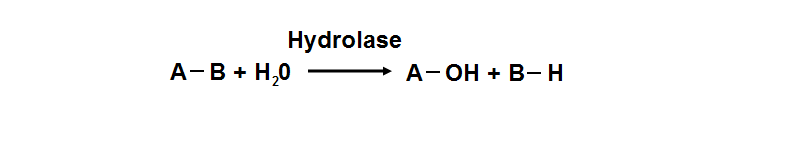
| * Hydrolase Activity
| | Hydrolyase catalyze the hydrosis of the chemical bond between A and B, resulting of 2 simple molecules |
|
| |
|
| * Magnesium Ion Binding
| |
|
| |
|
| * N-acylneuraminate-9-phosphatase Activity | | * Hydrolase |
| | ** Catalyze hydrolysis reaction |
| | ** Addition of the hydrogen and hydroxyl ions of water |
| | ** Splitting into 2 or more simpler molecules |
| | ** EC class 3 |
|
| |
|
| * Phosphoglycolate Phosphatase Activity
| |
|
| |
|
|
| |
|
| | == Function of sulfatases == |
|
| |
|
| | Sulfatases are enzymes,which hydrolyse sulfate ester bonds of substrates. |
| | Most of the family members has shown to contain a highly conserved cystine residue and a bivalent metal binding site. |
|
| |
|
|
| |
|
| [http://amigo.geneontology.org/cgi-bin/amigo/go.cgi?view=details&depth=1&query=0016787 1. Hydrolase Activity]
| |
|
| |
|
| [http://amigo.geneontology.org/cgi-bin/amigo/go.cgi?view=details&depth=1&query=0000287 2. Magnesium Ion Binding]
| |
|
| |
|
| [http://amigo.geneontology.org/cgi-bin/amigo/go.cgi?view=details&depth=1&query=0050124 3. N-acylneuraminate-9-phosphatase Activity]
| |
|
| |
|
| [http://amigo.geneontology.org/cgi-bin/amigo/go.cgi?view=details&depth=1&query=0008967 4. Phosphoglycolate Phosphatase Activity]
| | == Functional site== |
| | MSA data revealed some conserved residues on the sequence. They were mapped on the three dimantional structure. |
|
| |
|
| | | [[Image:Zn_Cl_surface]] |
| | |
| {| border="1" cellpadding="15" cellspacing="0"
| |
| |+'''Table of functions'''
| |
| |Catalytic activity
| |
| |N-acylneuraminate 9-phosphate + H2O = N-acylneuraminate + phosphate
| |
| |-
| |
| |Cofactor
| |
| |Magnesium (By similarity)
| |
| |-
| |
| |Enzyme regulation
| |
| |Inhibited by vanadate and calcium (By similarity)
| |
| |-
| |
| |Pathway
| |
| |Carbohydrate metabolism; aminosugar metabolism
| |
| |-
| |
| |Similarity
| |
| |Belongs to the haloacid dehalogenase-like hydrolase superfamily. NANP family
| |
| |}
| |
| | |
| | |
| Using ProFunc: Ligand-binding template search results for 2gfh.
| |
| | |
| Structural similarity: 91.5%
| |
| | |
| E-value < 1.00E-06 ( 7.22E-07)
| |
| | |
| Similarity score: 364.02
| |
| | |
| PBD id: 2hi0
| |
| | |
| Name: Hydrolase
| |
| | |
| Title: Crystal structure of putative phosphoglycolate phosphatase (yp_619066.1) from lactobacillus delbrueckii subsp. Bulgaricus atcc baa-365 at 1.51 a resolution
| |
| | |
| Source: Lactobacillus delbrueckii. Bacteria. Gene: yp_619066.1. Expressed in: escherichia coli.
| |
| | |
| Reaction: 2-phosphoglycolate + H2O = glycolate + phosphate
| |
| | |
| [[Image:Tree.JPG|thumb|Description|left]] | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| =='''GO Terms'''==
| |
|
| |
| Polymer: haloacid dehalogenase-like hydrolase domain containing 4
| |
| | |
| Molecular Function: None
| |
| | |
| Biological Process: None
| |
| | |
| Cellular Component: None
| |
| | |
| | |
| =='''Function in Human'''==
| |
| | |
| haloacid dehalogenase-like hydrolase domain
| |
| | |
| (Found in OMIM)
| |
| | |
| Neu5Ac-9-PHOSPHATE PHOSPHATASE HALOACID DEHALOGENASE-LIKE HYDROLASE DOMAIN-CONTAINING PROTEIN-4; HDHD4
| |
| Gene map locus 20p11
| |
| | |
| | |
| Neu5Ac-9-phosphate phosphatase (Neu5Ac-9-Pase; EC 3.1.3.29) dephosphorylates Neu5Ac-9-phosphate to form N-acetylneuraminate (Neu5Ac), the main form of sialic acid in vertebrates that has important roles in protein-protein and cell-cell recognition.
| |
| | |
| | |
| CLONING
| |
| | |
| Maliekal et al. (2006) purified Neu5Ac-9-Pase from rat liver and isolated the phosphatase activity. Using SDS-PAGE analysis and tandem mass spectrometry, they identified a haloacid dehalogenase-like hydrolase domain containing-4 (HDHD4) protein. Purified recombinant human HDHD4 catalyzed the dephosphorylation of Neu5Ac-9-phosphate with a catalytic efficiency more than 2 orders of magnitude higher than for any other substrate tested. The 248-amino acid human HDHD4 protein has 3 motifs found in phosphatases of the haloacid dehalogenase (HAD) family: the first with 2 extremely conserved aspartates, the second comprising a conserved serine or threonine, and the third comprising a conserved lysine and 2 conserved aspartates.
| |
| | |
| | |
| GENE STRUCTURE
| |
| | |
| Maliekal et al. (2006) determined that the human HDHD4 gene contains 2 exons.
| |
| | |
| | |
| GENE FUNCTION
| |
| | |
| Maliekal et al. (2006) determined that the phosphatase activity of human Neu5Ac-9-Pase protein was dependent on the presence of Mg(2+) and was inhibited by vanadate and Ca(2+), which is characteristic of members of the HAD family of phosphatases.
| |
| | |
| | |
| | |
| | |
| | |
| | |
| | |
| ----
| |
| | |
| info
| |
| | |
| N-acetylneuraminic acid phosphatase
| |
| | |
| Homologous to mouse (Mus musculus)
| |
| | |
| Haloacid Dehalogenase Like Hydrolase Domain Containing 4
| |
| | |
| Classified as Hydrolase
| |
| | |
| [http://www.ebi.ac.uk/thornton-srv/databases/profunc/index.html Infomation on ProFunc] (useful 2gfhA)
| |
| | |
| [http://www.ebi.ac.uk/pdbsum/ Information on PDBsum] (2gfh)
| |
| | |
| From [http://www.ebi.ac.uk/cgi-bin/sumtab?tool=fasta&jobid=fasta-20070515-07270763 FASTA]
| |
| most likely function is N-acetylneuraminic acid phosphatase.
| |
| | |
| The haloalkanoate dehalogenase superfamily (HADSF) is one of the largest and most ubiquitous enzyme families identified to date, with over 3,000 members in organisms ranging from bacteria to humans. Remarkable diversity of chemistry and function has emerged through evolution of the HAD catalytic scaffold. Despite the name, the dehalogenases, which catalyze carbon group transfer, represent a minute fraction of the family. All other known catalytic activities are directed at phosphoryl transfer. Numerous proteins from the HADSF are found in each organism (29 in E. coli and 58 in humans, for example) where they perform a diverse collection of novel physiological functions in primary and secondary metabolism, membrane transport, signal transduction, and nucleic acid repair. http://biophysics.bumc.bu.edu/faculty/allen/allenpage/had.htm
| |
| | |
| Haloacid dehalogenases (E.C.3.8.1.2) are members of the haloacid dehalogenase superfamily, which also contains ATPases, phosphatases and epoxide hydrolases. They catalyse the conversion of α-halo-carboxylic acids to the corresponding hydroxyalkanoic acid by nucleophilic attack on the α-carbon by a conserved aspartic acid residue to form an ester intermediate, which is then further hydrolysed by a water molecule. There are three subtypes of haloacid dehalogenase based on substrate specificity; those that can use both enantiomers as substrates, those that act only on the L enantiomer and those that act only on the D enantiomer.
| |
|
| |
|
| ---- | | ---- |
|
| |
|
| Press here to go [http://compbio.chemistry.uq.edu.au/mediawiki/index.php/BIOL3004_2007 ''Back'']
| | Click here to go [http://compbio.chemistry.uq.edu.au/mediawiki/index.php/BIOL3004_2007 ''Back''] |