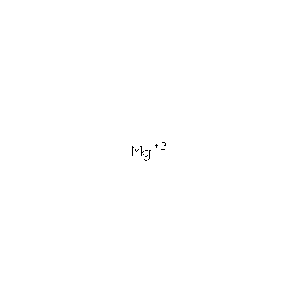Functional analysis: Difference between revisions
No edit summary |
No edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== '''Haloacid dehalogenase-like hydrolase domain containing 2''' (2HO4:A,B) == | |||
===Name of the protein=== | |||
====Hydrolase==== | |||
Haloacid dehalogenase(HAD) is the name of the compound superfamily. Hydrolase refers to the protein that has hydrolase activity i.e. the catalysing the hydrolysis of various bonds. The breakdown process, hydrolysis, is a replacing mechanism to the covalent bonding with water molecule. The product will become more water soluble and produce free enzyme.[http://amigo.geneontology.org/cgi-bin/amigo/go.cgi?view=details&depth=1&query=0016787 /Link] | |||
---- | |||
===Function are depend on the structure=== | |||
Use Mouse Genome Informatics(MGI)search engine, we found '''5 Category of Function or Process''' with evidence about this protein. Four molecular functions and one biological process are classified in this protein. They are catalytic activity, hydrolase activity, magnesium ion binding and metabolic process. The fundamental cellular process, the biological processess work together and compromise. | |||
GO Annotations in Tabular Form[http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=markerGO&key=60967 /Link] | |||
{| border="1" cellspacing="0" cellpadding="5" | |||
! Category | |||
! Classification | |||
! Term Evidence | |||
! Inferred From | |||
! Ref(s) | |||
|- | |||
| Molecular Function | |||
| catalytic activity | |||
| IEA | |||
| InterPro:IPR005834 | |||
| J:72247 | |||
|- | |||
| Molecular Function | |||
| hydrolase activity | |||
| IEA | |||
| InterPro:IPR006357 | |||
| J:72247 | |||
|- | |||
| Molecular Function | |||
| hydrolase activity | |||
| IEA | |||
| SP_KW:KW-0378 | |||
| J:60000 | |||
|- | |||
| Molecular Function | |||
| magnesium ion binding | |||
| IEA | |||
| SP_KW:KW-0460 | |||
| J:60000 | |||
|- | |||
| Biological Process | |||
| metabolic process | |||
| IEA | |||
| InterPro:IPR005834,InterPro:IPR006357 | |||
| J:72247 | |||
|- | |||
|} | |||
<pre> | |||
Gene Ontology Evidence Code Abbreviations: | |||
IC Inferred by curator | |||
IDA Inferred from direct assay | |||
IEA Inferred from electronic annotation | |||
IGI Inferred from genetic interaction | |||
IMP Inferred from mutant phenotype | |||
IPI Inferred from physical interaction | |||
ISS Inferred from sequence or structural similarity | |||
NAS Non-traceable author statement | |||
ND No biological data available | |||
RCA Reviewed computational analysis | |||
TAS Traceable author statement | |||
</pre> | |||
The evidence was obtained by Mammalian Orthology Load, and the information was derived from Homologene National Center for Biotechnology Information. This protein was reviewed in 2004 and record with J Number: J:90500. The Mammalian Orthology Query Result proven, the evidence was from using both '''Amino acid sequence comparison''' and '''Nucleotide sequence comparison''' on different species. And the results shown '''human, mouse, rat, chimpanzee and dog''' all have the sequence of this protein.[http://www.informatics.jax.org/searches/homology_report.cgi?_Marker_key=60967 /Link] | |||
---- | |||
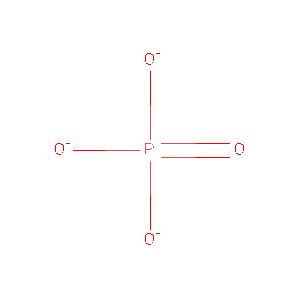
===Ligands and Prosthetic Groups=== | |||
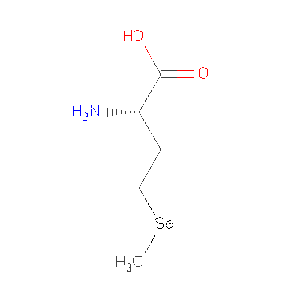
Protein Data Bank (PDB) summaries the structure, biological and chemistry and sequence detail of the protein. Under these reports, the PBD Biology and Chemistry Report shows the protein contain following Ligands and Prosthetic Groups; Phosphate Ion, Magnesium Ion and Selenomethionine. | |||
''' Ligands and Prosthetic Groups''' | |||
{| border="1" cellspacing="0" cellpadding="5" | |||
! ID | |||
! Name Chemical | |||
! Formula | |||
! Weight Ligand | |||
! Link | |||
|- | |||
| PO4 | |||
| Phosphate Ion | |||
| O4 P 3- | |||
| 94.971 | |||
| [http://www.rcsb.org/pdb/ligand/ligandsummary.do?handler=biologyChemistry&hetId=PO4 /View] | |||
|- | |||
| '''MG''' | |||
| '''Magnesium Ion''' | |||
| '''Mg 2+ ''' | |||
| 24.305 | |||
| [http://www.rcsb.org/pdb/ligand/ligandsummary.do?handler=biologyChemistry&hetId=MG /View] | |||
|- | |||
| MSE | |||
| Selenomethionine | |||
| C5 H11 N O2 Se | |||
| 196.107 | |||
| [http://www.rcsb.org/pdb/ligand/ligandsummary.do?handler=biologyChemistry&hetId=MSE /View] | |||
|- | |||
|} | |||
''PBD Biology and Chemistry Report'' [http://www.rcsb.org/pdb/explore/biologyAndChemistry.do?structureId=2HO4 /''Link''] | |||
---- | |||
===Expression of the protein in the body=== | |||
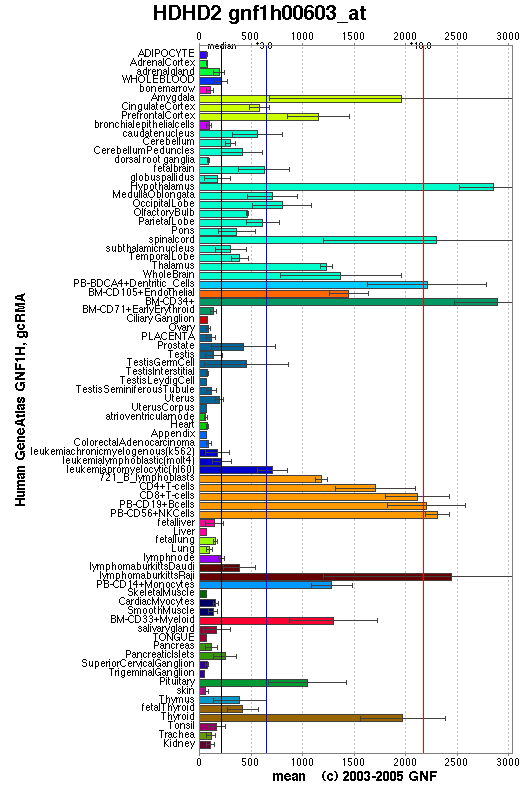
Genomics Institute of the Novartis Research Foundation summaries detail about protein. In the diagram below, haloacid dehalogenase-like hydrolase domain containing 2 sequence was calculated in different organisms of the body. From the result we can suggest this protein mainly functioning in '''Central Nerve System''' and '''Immune System'''. [http://symatlas.gnf.org/SymAtlas/symquery?q=212508 /Link] | |||
[[Image:HDHD2_at.png]] | |||
''The diagram illustrate the expression of protein or gene in different organisms. The sequence expression of the protein appears more in amygdala, cortex, hypothalamus, spinal cord, dentritic cells, T cells, B cells and lymphatic system. '' | |||
---- | |||
===Protein function=== | |||
====Basis of Haloacid dehalogenase family==== | |||
Identification of the Haloacid dehalogenase family on the basis of the structural similarities has been described in 1999. The HAD superfamily has similar reaction mechanism base on their structure. Members in this family include enzymes (such as L-2-haloacid dehalogenase, epoxide hydrolase), variety of phosphatases (e.g. phosphoserine phosphatase, phosphomannomutase, phosphoglycolate phosphatase and sucrose-phosphate synthase), and the catalytic subunits (such as the P-type ATPases). The ATPases are essential for the transport of cations across biological membranes, because it contains the binding sites for ATP and Mg2, and a phosphorylation site. ([http://www.biochemj.org/bj/339/0223/bj3390223.htm / Ridder et al.,1999]) | |||
[[Image:Protein2.JPG]] | |||
<pre> | |||
1. The mechanism starts with the binding of the phosphorylated substrate | |||
mediate by Mg2 ion. (In this diagram, the substrate is a serine residue) | |||
Followed by a nucleophilic attack by one of the carboxylate oxygen atoms and | |||
aspartate residue to form an acyl-phosphate intermediate. In this point, the | |||
positive charge of Mg2 ion is acting on phosphorus atom to stabilize it. | |||
2. The acyl-phosphate intermediate is hydrolysed by nucleophilic attack | |||
of a water molecule. | |||
3. In agreement of formation of an oxyanion intermediate on the aspartate residue, | |||
mediate by counterbalanced of the Mg2 ion, and hydrolase activity on | |||
phosphorylation site, this yield phosphate and free enzyme (serine residue). | |||
</pre> | |||
====Functional change of the protein==== | |||
A research was done in 1979, by administrating chlorpromazine at the rate of 15 milligram per kilogram to rats. By determining the mitochondria of the sensomotor cortex and localization of Mg-activated ATPase in the cortex, the result showed hydrolase activity increased in the brain after three hours of administration. The discussion suggested that the '''increase of ATP-hydrolase activity''' in the cortex will '''induce depression of glycolytic and respiratory activity of the cells'''. And this is due to increase of permeability of the mitochondrial membranes. ([http://www.springerlink.com/content/mmt8t16q20131540/ Kleshchinov et al., 1979]) | |||
---- | |||
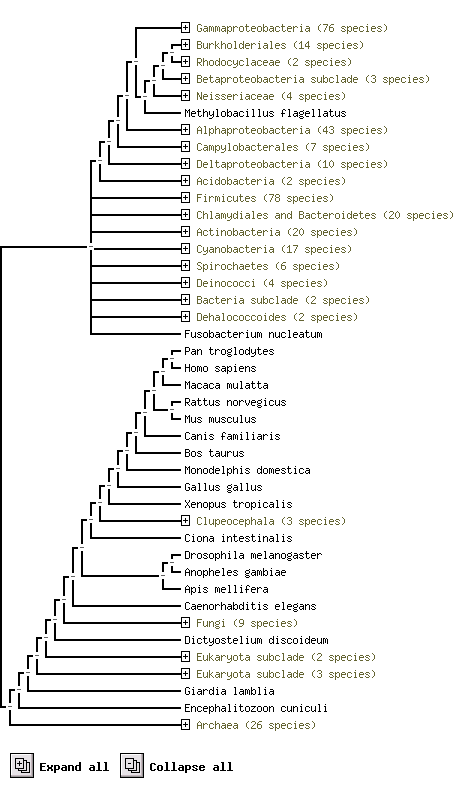
===What about it's functional partners=== | |||
From STRING website, by using the orthology information from the COG database, we predicted it's functional partners. KIAA0423 annotation is it best suit predicted Functional Partners, but it is not yet available. [http://string.embl.de/newstring_cgi/show_ajax_phylo_evidence.pl?taskId=NFsMuRjilfWk&allnodes=1 /Link] | |||
[[Image:HDHD2_and_predicted_functional_partner.png]] | |||
''The above result shows using protein sequence to compare difference species and their. e.g. Pan troglodytes and Homo sapiens are two species highly conservatity'' | |||
---- | |||
[[Image:PO4_400.gif|thumb|PO4]] | |||
[[Image:MG_400.gif|thumb|MG]] | |||
[[Image:MSE 400.gif|thumb|MSE]] | |||
---- | ---- | ||
[http://compbio.chemistry.uq.edu.au/mediawiki/index.php/ | [http://compbio.chemistry.uq.edu.au/mediawiki/index.php/Report:_Characterization_of_the_2HO4_protein_using_bioinformatics_tools | Back to report menu] | ||
Latest revision as of 23:22, 11 June 2007
Haloacid dehalogenase-like hydrolase domain containing 2 (2HO4:A,B)
Name of the protein
Hydrolase
Haloacid dehalogenase(HAD) is the name of the compound superfamily. Hydrolase refers to the protein that has hydrolase activity i.e. the catalysing the hydrolysis of various bonds. The breakdown process, hydrolysis, is a replacing mechanism to the covalent bonding with water molecule. The product will become more water soluble and produce free enzyme./Link
Function are depend on the structure
Use Mouse Genome Informatics(MGI)search engine, we found 5 Category of Function or Process with evidence about this protein. Four molecular functions and one biological process are classified in this protein. They are catalytic activity, hydrolase activity, magnesium ion binding and metabolic process. The fundamental cellular process, the biological processess work together and compromise.
GO Annotations in Tabular Form/Link
| Category | Classification | Term Evidence | Inferred From | Ref(s) |
|---|---|---|---|---|
| Molecular Function | catalytic activity | IEA | InterPro:IPR005834 | J:72247 |
| Molecular Function | hydrolase activity | IEA | InterPro:IPR006357 | J:72247 |
| Molecular Function | hydrolase activity | IEA | SP_KW:KW-0378 | J:60000 |
| Molecular Function | magnesium ion binding | IEA | SP_KW:KW-0460 | J:60000 |
| Biological Process | metabolic process | IEA | InterPro:IPR005834,InterPro:IPR006357 | J:72247 |
Gene Ontology Evidence Code Abbreviations: IC Inferred by curator IDA Inferred from direct assay IEA Inferred from electronic annotation IGI Inferred from genetic interaction IMP Inferred from mutant phenotype IPI Inferred from physical interaction ISS Inferred from sequence or structural similarity NAS Non-traceable author statement ND No biological data available RCA Reviewed computational analysis TAS Traceable author statement
The evidence was obtained by Mammalian Orthology Load, and the information was derived from Homologene National Center for Biotechnology Information. This protein was reviewed in 2004 and record with J Number: J:90500. The Mammalian Orthology Query Result proven, the evidence was from using both Amino acid sequence comparison and Nucleotide sequence comparison on different species. And the results shown human, mouse, rat, chimpanzee and dog all have the sequence of this protein./Link
Ligands and Prosthetic Groups
Protein Data Bank (PDB) summaries the structure, biological and chemistry and sequence detail of the protein. Under these reports, the PBD Biology and Chemistry Report shows the protein contain following Ligands and Prosthetic Groups; Phosphate Ion, Magnesium Ion and Selenomethionine.
Ligands and Prosthetic Groups
| ID | Name Chemical | Formula | Weight Ligand | Link |
|---|---|---|---|---|
| PO4 | Phosphate Ion | O4 P 3- | 94.971 | /View |
| MG | Magnesium Ion | Mg 2+ | 24.305 | /View |
| MSE | Selenomethionine | C5 H11 N O2 Se | 196.107 | /View |
PBD Biology and Chemistry Report /Link
Expression of the protein in the body
Genomics Institute of the Novartis Research Foundation summaries detail about protein. In the diagram below, haloacid dehalogenase-like hydrolase domain containing 2 sequence was calculated in different organisms of the body. From the result we can suggest this protein mainly functioning in Central Nerve System and Immune System. /Link
The diagram illustrate the expression of protein or gene in different organisms. The sequence expression of the protein appears more in amygdala, cortex, hypothalamus, spinal cord, dentritic cells, T cells, B cells and lymphatic system.
Protein function
Basis of Haloacid dehalogenase family
Identification of the Haloacid dehalogenase family on the basis of the structural similarities has been described in 1999. The HAD superfamily has similar reaction mechanism base on their structure. Members in this family include enzymes (such as L-2-haloacid dehalogenase, epoxide hydrolase), variety of phosphatases (e.g. phosphoserine phosphatase, phosphomannomutase, phosphoglycolate phosphatase and sucrose-phosphate synthase), and the catalytic subunits (such as the P-type ATPases). The ATPases are essential for the transport of cations across biological membranes, because it contains the binding sites for ATP and Mg2, and a phosphorylation site. (/ Ridder et al.,1999)
1. The mechanism starts with the binding of the phosphorylated substrate mediate by Mg2 ion. (In this diagram, the substrate is a serine residue) Followed by a nucleophilic attack by one of the carboxylate oxygen atoms and aspartate residue to form an acyl-phosphate intermediate. In this point, the positive charge of Mg2 ion is acting on phosphorus atom to stabilize it. 2. The acyl-phosphate intermediate is hydrolysed by nucleophilic attack of a water molecule. 3. In agreement of formation of an oxyanion intermediate on the aspartate residue, mediate by counterbalanced of the Mg2 ion, and hydrolase activity on phosphorylation site, this yield phosphate and free enzyme (serine residue).
Functional change of the protein
A research was done in 1979, by administrating chlorpromazine at the rate of 15 milligram per kilogram to rats. By determining the mitochondria of the sensomotor cortex and localization of Mg-activated ATPase in the cortex, the result showed hydrolase activity increased in the brain after three hours of administration. The discussion suggested that the increase of ATP-hydrolase activity in the cortex will induce depression of glycolytic and respiratory activity of the cells. And this is due to increase of permeability of the mitochondrial membranes. (Kleshchinov et al., 1979)
What about it's functional partners
From STRING website, by using the orthology information from the COG database, we predicted it's functional partners. KIAA0423 annotation is it best suit predicted Functional Partners, but it is not yet available. /Link
The above result shows using protein sequence to compare difference species and their. e.g. Pan troglodytes and Homo sapiens are two species highly conservatity