Results 2ece: Difference between revisions
No edit summary |
|||
| Line 385: | Line 385: | ||
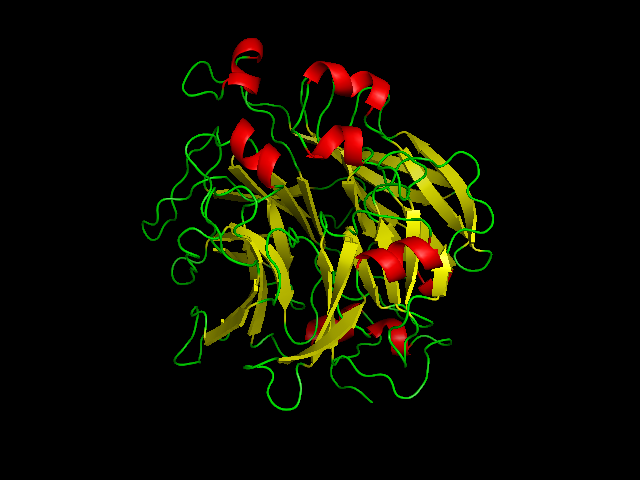
== FUNCTIONAL analysis of SELENBP 1 (pdb 2ece) == | |||
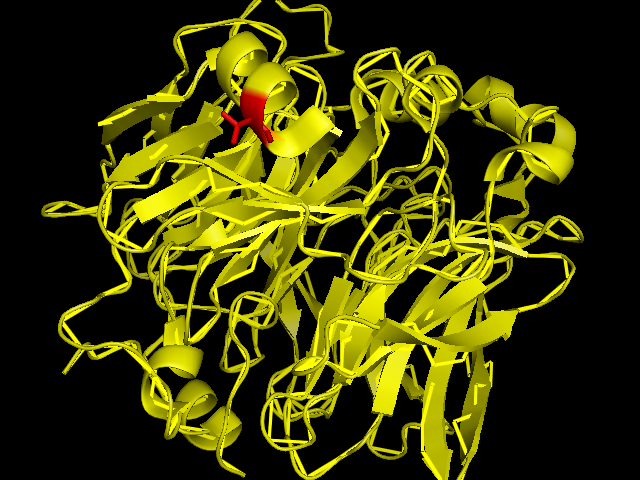
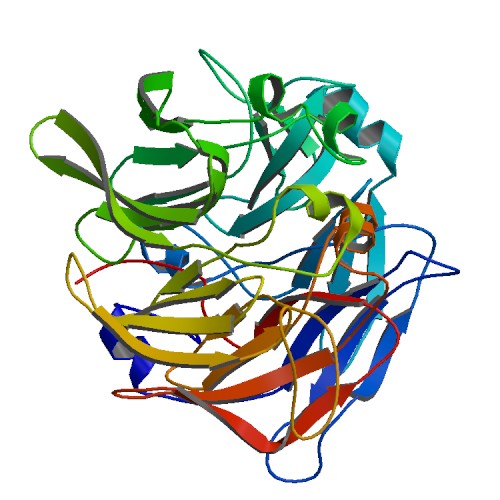
The red coloured region is the region carring Se, and it is evident that the Se is incorporated in an amino acid of the helix chain.(Pymol Image) | [[Image:SBP with Sele atom.png|frame|border|left|The red coloured region is the region carring Se, and it is evident that the Se is incorporated in an amino acid of the helix chain.(Pymol Image) | ||
]] | |||
Revision as of 11:05, 9 June 2008
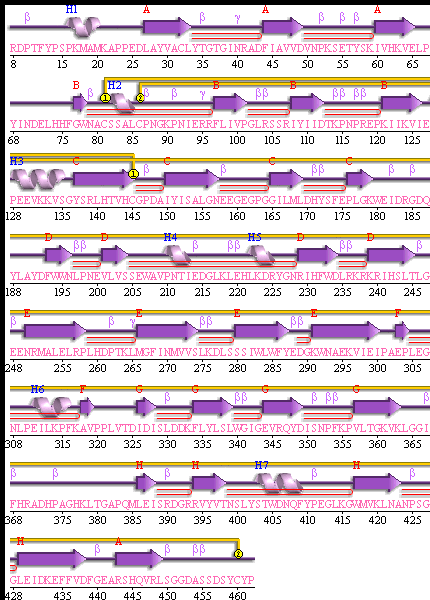
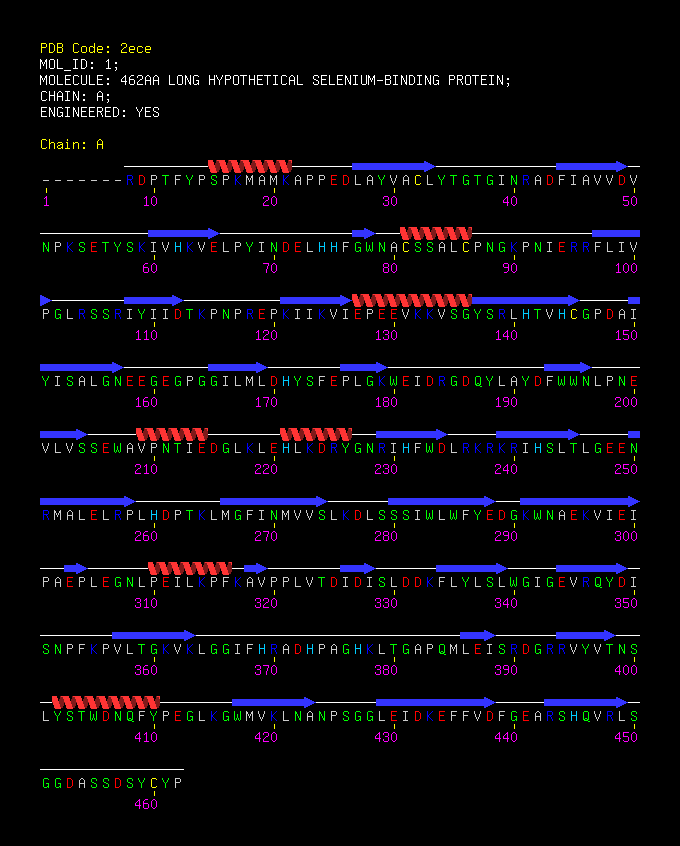
STRUCTURAL ANALYSIS
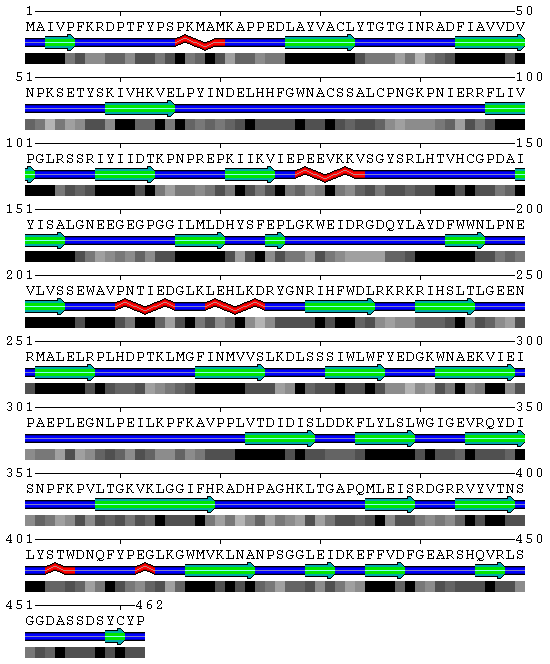
SBP 1 Amino Acid FASTA FORMAT Sequence
>2ECE:A|PDBID|CHAIN|SEQUENCE
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKIVHKVELPYINDELHHFGWNA CSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREPKIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEG EGPGGILMLDHYSFEPLGKWEIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEIPAEPLEGNLPEILKPFKAVP PLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTGKVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNS LYSTWDNQFYPEGLKGWMVKLNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP
WARNING! Given sequence appeared to be a soluble protein, no TM domains found!
Output format is the following:
1st line -> residue numeration
2nd line -> query amino acid sequence
3rd line -> trans-membrane domain prediction (T-TM region, N-soluble part)
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKI
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
VHKVELPYINDELHHFGWNACSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREP
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
KIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEGEGPGGILMLDHYSFEPLGKW
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
EIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEI
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
PAEPLEGNLPEILKPFKAVPPLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTG
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
KVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNSLYSTWDNQFYPEGLKGWMVK
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
LNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
Figure 3 SABLE server results.
STRUCTURAL COMPARISONS
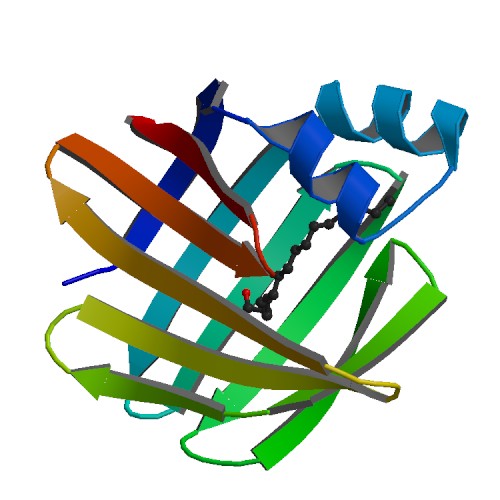
Explore SBP features and structural summary here [3].The domains of SBP are shown here [4]
Notice how the domains are similar to the putative Isomerase domains of E.coli on Figure 10below.
1RI6 DOMAINS
2ECE DOMAINS