FUNCTIONAL ANALYSIS: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
[[Literature supporting conclusion]] | [[Literature supporting conclusion]] | ||
From a summary of predicted function from our [http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/profunc/GetResults.pl?source=profunc&user_id=ay99&code=053949 ProFunc results] we can see that the most commonly occurring name term is GTPase, followed by GTP binding. | |||
During the ProFunc analysis [http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/profunc/runprog.pl?prog=funchtml&html_name=seq001_aln.html&pdbcode=ay99&user_id=&pdb_type=PROFUNC&code=053949 UniProt] was used to perform a multiple sequence alignment on our protein sequence. The top 2 results were both highly conserved as well as also being YlqF proteins. The rest of the 10 proteins listed were all either GTPase or GTP binding proteins. This sequence search supports the literature in its claim that the YlqF protein is required for the late step of 50s ribosomal subunit assembly. | During the ProFunc analysis [http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/profunc/runprog.pl?prog=funchtml&html_name=seq001_aln.html&pdbcode=ay99&user_id=&pdb_type=PROFUNC&code=053949 UniProt] was used to perform a multiple sequence alignment on our protein sequence. The top 2 results were both highly conserved as well as also being YlqF proteins. The rest of the 10 proteins listed were all either GTPase or GTP binding proteins. This sequence search supports the literature in its claim that the YlqF protein is required for the late step of 50s ribosomal subunit assembly. | ||
| Line 21: | Line 24: | ||
Q03KX8 47.6 - Q03KX8 Predicted GTPase.</pre> | Q03KX8 47.6 - Q03KX8 Predicted GTPase.</pre> | ||
Revision as of 14:33, 10 June 2007
50S ribosome assembly in Bacillus subtilis
Literature supporting conclusion
From a summary of predicted function from our ProFunc results we can see that the most commonly occurring name term is GTPase, followed by GTP binding.
During the ProFunc analysis UniProt was used to perform a multiple sequence alignment on our protein sequence. The top 2 results were both highly conserved as well as also being YlqF proteins. The rest of the 10 proteins listed were all either GTPase or GTP binding proteins. This sequence search supports the literature in its claim that the YlqF protein is required for the late step of 50s ribosomal subunit assembly.
Aligned sequences:
%-tage
Seq. id. identity Name
-------- -------- ----
Query - - Query sequence
O31743 95.6 - O31743 YlqF protein.
Q65JP4 84.6 - Q65JP4 YlqF (GTP-binding domain protein).
Q845L2 70.1 - Q845L2 Hypothetical protein.
Q9Z9S1 59.9 - Q9Z9S1 YlqF (BH2476 protein).
Q4MFY1 65.9 - Q4MFY1 Hypothetical protein.
Q5WFP0 53.5 - Q5WFP0 GTPase.
Q5HPU7 50.9 - Q5HPU7 GTP-binding protein, putative.
Q3XZB6 52.4 - Q3XZB6 GTP-binding protein.
A0Q0X8 49.3 - A0Q0X8 GTP-binding protein, putative.
Q03KX8 47.6 - Q03KX8 Predicted GTPase.
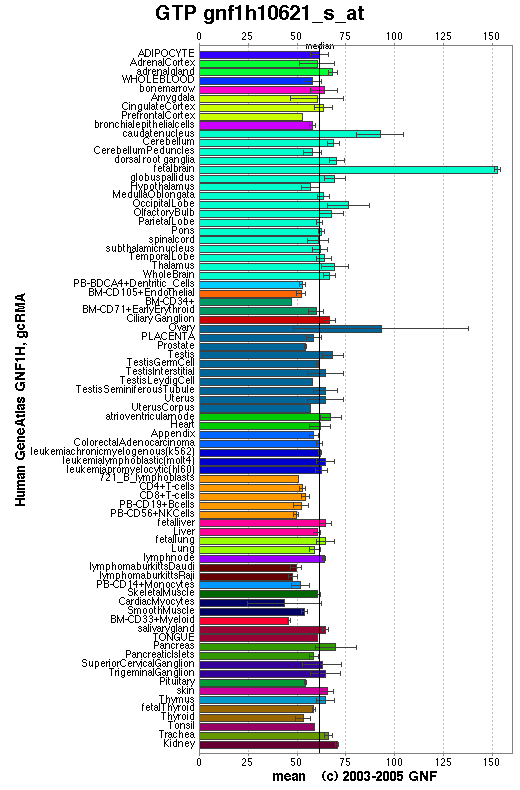
From the distribution of the human ortholog protein in the human body it is evident that this protein is widely distributed and therefore commonly used. These results are consistent with our claim that 1pujA is a GTPase.
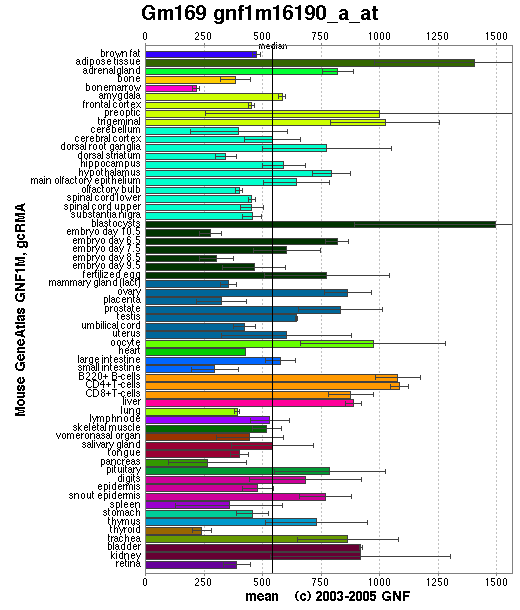
Though the distribution of the mouse ortholog protein is somewhat more scattered than the human ortholog protein it is still relatively common in all major tissues meaning that this is also consistent with our claim that 1pujA is a GTPase.
GO Code.Bayesian Score.Evidence Rank.Number of Clues.Description
Molecular Function
0005525 0.9815 2.8 6 GTP binding
0000166 0.0101 2.9 6 nucleotide binding
0008168 0.0068 2.9 6 methyltransferase activity
0003924 0.0016 2.5 6 GTPase activity
Biological Process
0007264 1.0000 2.8 6 small GTPase mediated signal transduction
The ProKnow analysis shows that 1pujA's molecular function is GTP binding and its biological function is small GTPase mediated signal transduction.