Results 5: Difference between revisions
No edit summary |
mNo edit summary |
||
| Line 67: | Line 67: | ||
{| border="1" | {| border="1" | ||
|- | |- | ||
|Class | |'''Class''' | ||
Alpha and beta proteins | Alpha and beta proteins | ||
|Fold | |'''Fold''' | ||
P-loop containing | P-loop containing | ||
nucleoside triphosphate hydrolases | nucleoside triphosphate hydrolases | ||
|Superfamily | |'''Superfamily''' | ||
P-loop containing | P-loop containing | ||
nucleoside triphosphate hydrolases | nucleoside triphosphate hydrolases | ||
|Family | |'''Family''' | ||
G proteins | G proteins | ||
|Domain | |'''Domain''' | ||
Probable GTPase YlqF | Probable GTPase YlqF | ||
|Species | |'''Species''' | ||
Bacillus subtilis | Bacillus subtilis | ||
|- | |- | ||
Revision as of 04:39, 11 June 2007
STRUCTURE
Quality of YlqF protein model and overall structure
The asymmetric unit of Bacillus subtilis YlqF protein consists of a polymer containing 282 amino acids (Figure 1). The protein has been refined at 2 angstroms to a crystallographic R factor of 21.6% and free R factor of 25%. Table 1 summarizes the refinement statistics including protein quality parameters. MolProbity Ramachandran analysis of YlqF in shows that 96.5% of all residues lie in the favoured regions and 98.8% of all residues lie in the allowed regions.
Table 1. Crystal parameters and refinement statistics
| Parameters | Resolution[Å]
2.00 |
R factor, %
21.6 |
Free R factor, %
25.0 |
Space Group
P 21 21 21 |
| Unit Cell | Length[Å]
Angles [°] |
a
alpha |
36.75
90.00 |
b
beta |
68.57
90.00 |
c
gamma |
105.57
90.00 |
|---|
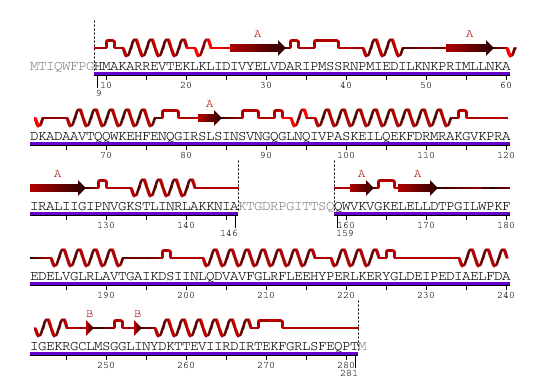
1 - MTIQWFPGHM AKARREVTEK LKLIDIVYEL VDARIPMSSR NPMIEDILKN KPRIMLLNKA 61 - DKADAAVTQQ WKEHFENQGI RSLSINSVNG QGLNQIVPAS KEILQEKFDR MRAKGVKPRA 121 - IRALIIGIPN VGKSTLINRL AKKNIAKTGD RPGITTSQQW VKVGKELELL DTPGILWPKF 181 - EDELVGLRLA VTGAIKDSII NLQDVAVFGL RFLEEHYPER LKERYGLDEI PEDIAELFDA 241 - IGEKRGCLMS GGLINYDKTT EVIIRDIRTE KFGRLSFEQP TM
Figure 1. Amino acid sequence of YlqF
The secondary structure of YlqF mainly contains 50% helical (13 helices; 142 residues) and 10% beta sheet (6 strands; 31 residues)(see Figure 2). YlqF protein consists of two domains. One domain contains Rossmann fold with α/β class. This domain possesses 1-177 residues, and forms a 3-layer sandwich structure. The other one is referred to as a conserved hypothetical protein with mainly α class. This possesses 178-282 residues, and forms a orthogonal bundle structure. YlqF is also classified as a signalling protein. The molecular weight of the protein is 31986 Da.
![]() = pi helix,
= pi helix, ![]() = 310 helix,
= 310 helix, ![]() = extended strand,
= extended strand, ![]() = turn,
= turn, ![]() = alpha helix,
= alpha helix,
Greyed out residues have no structural information
Figure 2. Sequence and Secondary Structure
Structure Analysis
Analysis of SCOP classified YlqF as shown in Table 2.
| Class
Alpha and beta proteins |
Fold
P-loop containing nucleoside triphosphate hydrolases |
Superfamily
P-loop containing nucleoside triphosphate hydrolases |
Family
G proteins |
Domain
Probable GTPase YlqF |
Species
Bacillus subtilis |