Results 5: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
==='''Quality of YlqF protein model and overall structure'''=== | ==='''Quality of YlqF protein model and overall structure'''=== | ||
The asymmetric unit of Bacillus subtilis YlqF protein consists of a polymer containing 282 amino acids (Figure 1). The protein has been refined at 2 angstroms to a crystallographic R factor of 21.6% and free R factor of 25%. Table 1 summarizes the refinement statistics including protein quality parameters. MolProbity Ramachandran analysis of YlqF | The asymmetric unit of Bacillus subtilis YlqF protein consists of a polymer containing 282 amino acids (Figure 1). The protein has been refined at 2 angstroms to a crystallographic R factor of 21.6% and free R factor of 25%. Table 1 summarizes the refinement statistics including protein quality parameters. MolProbity Ramachandran analysis of YlqF shows that 96.5% of all residues lie in the favoured regions and 98.8% of all residues lie in the allowed regions. | ||
Revision as of 05:31, 11 June 2007
STRUCTURE
Quality of YlqF protein model and overall structure
The asymmetric unit of Bacillus subtilis YlqF protein consists of a polymer containing 282 amino acids (Figure 1). The protein has been refined at 2 angstroms to a crystallographic R factor of 21.6% and free R factor of 25%. Table 1 summarizes the refinement statistics including protein quality parameters. MolProbity Ramachandran analysis of YlqF shows that 96.5% of all residues lie in the favoured regions and 98.8% of all residues lie in the allowed regions.
Table 1. Crystal parameters and refinement statistics
| Parameters | Resolution[Å]
2.00 |
R factor, %
21.6 |
Free R factor, %
25.0 |
Space Group
P 21 21 21 |
| Unit Cell | Length[Å]
Angles [°] |
a
alpha |
36.75
90.00 |
b
beta |
68.57
90.00 |
c
gamma |
105.57
90.00 |
|---|
1 - MTIQWFPGHM AKARREVTEK LKLIDIVYEL VDARIPMSSR NPMIEDILKN KPRIMLLNKA 61 - DKADAAVTQQ WKEHFENQGI RSLSINSVNG QGLNQIVPAS KEILQEKFDR MRAKGVKPRA 121 - IRALIIGIPN VGKSTLINRL AKKNIAKTGD RPGITTSQQW VKVGKELELL DTPGILWPKF 181 - EDELVGLRLA VTGAIKDSII NLQDVAVFGL RFLEEHYPER LKERYGLDEI PEDIAELFDA 241 - IGEKRGCLMS GGLINYDKTT EVIIRDIRTE KFGRLSFEQP TM
Figure 1. Amino acid sequence of YlqF
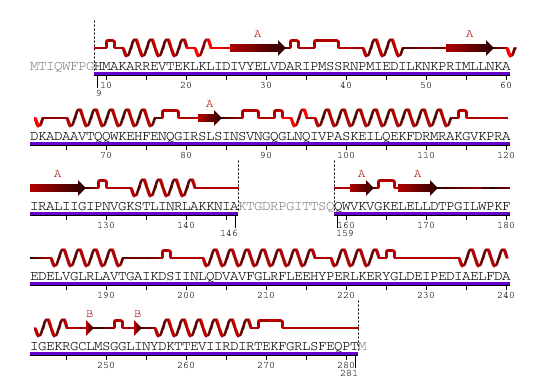
The secondary structure of YlqF mainly contains 50% helical (13 helices; 142 residues) and 10% beta sheet (6 strands; 31 residues)(see Figure 2). YlqF protein consists of two domains. One domain contains Rossmann fold with α/β class. This domain possesses 1-177 residues, and forms a 3-layer sandwich structure. The other one is referred to as a conserved hypothetical protein with mainly α class. This possesses 178-282 residues, and forms a orthogonal bundle structure. YlqF is also classified as a signalling protein. The molecular weight of the protein is 31986 Da.
![]() = pi helix,
= pi helix, ![]() = 310 helix,
= 310 helix, ![]() = extended strand,
= extended strand, ![]() = turn,
= turn, ![]() = alpha helix,
= alpha helix,
Greyed out residues have no structural information
Figure 2. Sequence and Secondary Structure
Structure Analysis
Structure classification of proteins (SCOP) classified YlqF as shown in Table 2. The results of Pfam classification described YlqF as GTPase of unknown function, Rhomboid family, and catalytic domain of alpha amylase.
| Class
Alpha and beta proteins |
Fold
P-loop containing nucleoside triphosphate hydrolases |
Superfamily
P-loop containing nucleoside triphosphate hydrolases |
Family
G proteins |
Domain
Probable GTPase YlqF |
Species
Bacillus subtilis |
No Chain raw-score Z-score %id lali rmsd Description 1 1pujA 3946.0 42.7 100 261 0.0 CONSERVED HYPOTHETICAL PROTEIN YLQF 2 1udxA 618.8 6.2 14 141 25.5 THE GTP-BINDING PROTEIN OBG 3 1uadA 541.8 6.1 9 99 2.6 RAS-RELATED PROTEIN RAL-A 4 1zbdA 553.7 6.1 12 99 2.8 RAB-3A 5 1g17A 526.0 5.9 18 95 2.5 RAS-RELATED PROTEIN SEC4 6 1c1yA 515.7 5.8 12 96 2.8 RAS-RELATED PROTEIN RAP-1A 7 1kjzA 571.3 5.7 14 114 4.0 EIF2GAMMA 8 1oixA 510.9 5.4 11 94 2.6 RAS-RELATED PROTEIN RAB-11A 9 1ukvY 519.2 5.3 10 99 2.8 SECRETORY PATHWAY GDP DISSOCIATION INHIBITOR 10 1ijfA 535.8 5.3 13 103 3.6 ELONGATION FACTOR 1-ALPHA 11 1gpmA 481.7 5.2 7 117 4.0 GMP SYNTHETASE 12 1hrkA 483.7 5.1 5 116 4.2 FERROCHELATASE 13 1d5cA 474.4 5.1 10 92 2.6 RAB6 GTPASE 14 1ni5A 512.4 5.0 6 115 5.5 PUTATIVE CELL CYCLE PROTEIN MESJ 15 3rapS 484.1 5.0 9 92 2.6 G PROTEIN RAP2A 16 1gwnA 475.5 5.0 10 94 2.7 RHO-RELATED GTP-BINDING PROTEIN RHOE 17 1vg8A 479.0 5.0 11 106 3.9 RAS-RELATED PROTEIN RAB-7 18 1cqxA 438.8 4.9 4 116 4.2 FLAVOHEMOPROTEIN 19 1mkyA 460.7 4.7 11 100 8.8 PROBABLE GTP-BINDING PROTEIN ENGA 20 1fzqA 463.8 4.7 14 92 2.6 ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 3
Table . Dali search results of PDB/chain identifiers and structural alignment statistics