Arylformamidase: Difference between revisions
Thomasparker (talk | contribs) |
Thomasparker (talk | contribs) No edit summary |
||
| Line 2: | Line 2: | ||
== Introduction == | == Introduction == | ||
why it was chosen for structural determination | |||
how its annotation arrived at 'arylformamidase | |||
information from pdb - structure details, etc... (perhaps picture) | |||
paragraph on silicibacter sp tm1040 - evolutionary info (where it fits in) (diagram) | |||
== Results == | == Results == | ||
most similar sequence - catalytic triad, structure with highlighted | |||
The most similar sequence with functional information available was that of an arylformamidase isolated from the liver of Mus musculus (see figure ...). A functional analysis of this protein has been performed identifying a catalytic triad using site-directed mutagenesis (Pabarcus et al. 2007). Conservation of this catalytic triad with 2pbl was assessed. Both residues S162 and H279 were found to be conserved in relatively conserved regions of the alignment. However, D247 had undergone a semi-conservative substitution. These residues correlated to S136, E214 and H241 of 2pbl which were subsequently located on the tertiary structure and determined to be sufficiently proximal to one another for catalysis (see figure...). | The most similar sequence with functional information available was that of an arylformamidase isolated from the liver of Mus musculus (see figure ...). A functional analysis of this protein has been performed identifying a catalytic triad using site-directed mutagenesis (Pabarcus et al. 2007). Conservation of this catalytic triad with 2pbl was assessed. Both residues S162 and H279 were found to be conserved in relatively conserved regions of the alignment. However, D247 had undergone a semi-conservative substitution. These residues correlated to S136, E214 and H241 of 2pbl which were subsequently located on the tertiary structure and determined to be sufficiently proximal to one another for catalysis (see figure...). | ||
[[Image:arylformamidase_alignment.png|centre|framed|'''Figure 3:''' ''Conservation of the catalytic triad between Arylformamidase and 2pbl.'']] | [[Image:arylformamidase_alignment.png|centre|framed|'''Figure 3:''' ''Conservation of the catalytic triad between Arylformamidase and 2pbl.'']] | ||
most similar structure - catalytic triad, structure with highlighted | |||
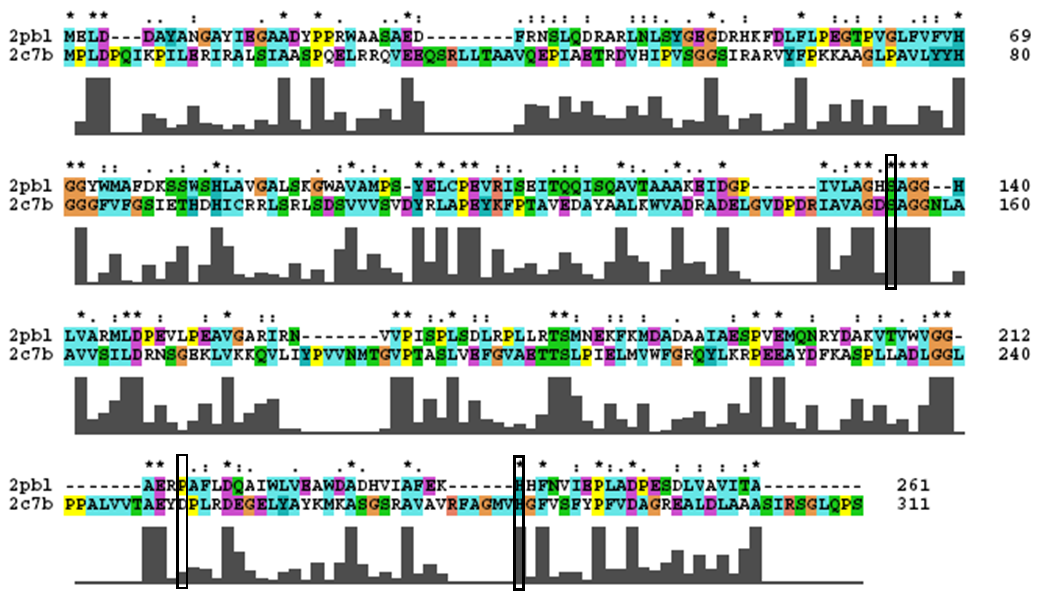
2pbl was found to share most structural similarity with a thermostable carboxylesterase from an uncultured archaeon (PDB ID: 2c7b; see figure ...). 2c7b shares a 16% sequence identity with 2pbl. From its structure, a catalytic triad has been identified (how?). To substantiate any functional similarity between 2pbl and 2c7b, conservation of the 2c7b catalytic triad was analysed (see figure ...). All three residues were found to be conserved, though H... and E... were found to match is less conserved regions. | 2pbl was found to share most structural similarity with a thermostable carboxylesterase from an uncultured archaeon (PDB ID: 2c7b; see figure ...). 2c7b shares a 16% sequence identity with 2pbl. From its structure, a catalytic triad has been identified (how?). To substantiate any functional similarity between 2pbl and 2c7b, conservation of the 2c7b catalytic triad was analysed (see figure ...). All three residues were found to be conserved, though H... and E... were found to match is less conserved regions. | ||
[[Image:2c7b_alignment.png|centre|framed|'''Figure 1:''' ''Conservation of the catalytic triad between 2cb7 and 2pbl.'']] | [[Image:2c7b_alignment.png|centre|framed|'''Figure 1:''' ''Conservation of the catalytic triad between 2cb7 and 2pbl.'']] | ||
presentation of tree and alignments | |||
== Discussion == | == Discussion == | ||
probable function | |||
The function of 2c7b has been well characterised (references). | |||
additional info on probable function | |||
Carboxylesterases have a common reaction mechanism (see figure ...). This is somewhat similar to the arylformamidase reaction mechanism incorporating hydrolysis of a ... bond. | Carboxylesterases have a common reaction mechanism (see figure ...). This is somewhat similar to the arylformamidase reaction mechanism incorporating hydrolysis of a ... bond. | ||
evolutionary link... | |||
biological implications/applications... | |||
Potential thermostability - applicability in industry. | Potential thermostability - applicability in industry. | ||
further research... | |||
== Conclusion == | == Conclusion == | ||
Revision as of 11:28, 31 May 2008
Abstract
Introduction
why it was chosen for structural determination
how its annotation arrived at 'arylformamidase
information from pdb - structure details, etc... (perhaps picture)
paragraph on silicibacter sp tm1040 - evolutionary info (where it fits in) (diagram)
Results
most similar sequence - catalytic triad, structure with highlighted
The most similar sequence with functional information available was that of an arylformamidase isolated from the liver of Mus musculus (see figure ...). A functional analysis of this protein has been performed identifying a catalytic triad using site-directed mutagenesis (Pabarcus et al. 2007). Conservation of this catalytic triad with 2pbl was assessed. Both residues S162 and H279 were found to be conserved in relatively conserved regions of the alignment. However, D247 had undergone a semi-conservative substitution. These residues correlated to S136, E214 and H241 of 2pbl which were subsequently located on the tertiary structure and determined to be sufficiently proximal to one another for catalysis (see figure...).
most similar structure - catalytic triad, structure with highlighted
2pbl was found to share most structural similarity with a thermostable carboxylesterase from an uncultured archaeon (PDB ID: 2c7b; see figure ...). 2c7b shares a 16% sequence identity with 2pbl. From its structure, a catalytic triad has been identified (how?). To substantiate any functional similarity between 2pbl and 2c7b, conservation of the 2c7b catalytic triad was analysed (see figure ...). All three residues were found to be conserved, though H... and E... were found to match is less conserved regions.
presentation of tree and alignments
Discussion
probable function
The function of 2c7b has been well characterised (references).
additional info on probable function
Carboxylesterases have a common reaction mechanism (see figure ...). This is somewhat similar to the arylformamidase reaction mechanism incorporating hydrolysis of a ... bond.
evolutionary link...
biological implications/applications...
Potential thermostability - applicability in industry.
further research...
Conclusion
Methods
Literature search
A literature search was performed using the putative annotation ‘arylformamidase’. A paper by Pabarcus et al. 2007 was returned which, ironically, described the arylformamidase from Mus Musculus.
Conservation of Catalytic Triad
An alignment of 2pbl and the protein of interest was performed using ClustalX. Default parameters were used. Residues of the catalytic triad were identified from the paper describing it and located in the sequence. Conservation of the residue and the surrounding sequence was observed. Note: in analysing conservation of the 2c7b catalytic triad with 2pbl, the clustalW alignment was found to differ from the alignment provided as part of the DALI results.
Additional Materials
Presentations
Sequence & Homology - Sebastian Mynott
Structure - Basma Al Alaiwat
Function - Thomas Parker
Links
Protein Data Bank Entry for 2PBL
FASTA Sequence
>gi|146387357|pdb|2PBL|A Chain A, Crystal Structure Of Putative Thioesterase (Yp_614486.1) From Silicibacter Sp. Tm1040 At 1.79 A Resolution GXELDDAYANGAYIEGAADYPPRWAASAEDFRNSLQDRARLNLSYGEGDRHKFDLFLPEGTPVGLFVFVH GGYWXAFDKSSWSHLAVGALSKGWAVAXPSYELCPEVRISEITQQISQAVTAAAKEIDGPIVLAGHSAGG HLVARXLDPEVLPEAVGARIRNVVPISPLSDLRPLLRTSXNEKFKXDADAAIAESPVEXQNRYDAKVTVW VGGAERPAFLDQAIWLVEAWDADHVIAFEKHHFNVIEPLADPESDLVAVITA
References
Pabarcus MK, Casida JE. Cloning, expression, and catalytic triad of recombinant arylformamidase. Protein Expr Purif. Nov;44(1):39-44.
De Simone G, Menchise V, Manco G, Mandrich L, Sorrentino N, Lang D, Rossi M, Pedone C. The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus. J Mol Biol. 2001 Nov 30;314(3):507-18.
Byun JS, Rhee JK, Kim ND, Yoon J, Kim DU, Koh E, Oh JW, Cho HS. Crystal structure of hyperthermophilic esterase EstE1 and the relationship between its dimerization and thermostability properties. BMC Struct Biol. 2007 Jul 12;7:47.
2-Effect of arylformamidase (kynurenine formamidase) gene inactivation in mice on enzymatic activity, kynurenine pathway metabolites and phenotype
http://www.ncbi.nlm.nih.gov/pubmed/12007602?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DiscoveryPanel.Pubmed_Discovery_RA&linkpos=2&log$=relatedarticles&dbfrom=pubmed