3b5q Structure and Function: Difference between revisions
No edit summary |
No edit summary |
||
| Line 6: | Line 6: | ||
[[Image:reaction.png]] | |||
[[Image:hydrolase.png]] | [[Image:hydrolase.png]] | ||
Revision as of 11:51, 6 June 2008
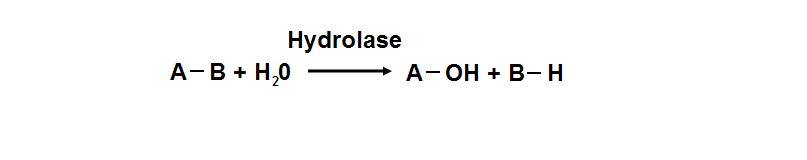
Protein data base profile characteris arylsulfatase as a hydrolase and a sulfatase. A hydrolase is an enzyme which hydrolyses a chemical bond. The general reaction can be shown as follows.
Hydrolases are named as EC 3 in enzyme classification.
Function of sulfatases
Sulfatases are enzymes,which hydrolyse sulfate ester bonds of substrates. These are categorised as EC 3.1.6. in enzyme classifications. Most of the family members has shown to contain a highly conserved cystine residue and a bivalent metal binding site.
Arylsulphatase A (ASA) is a lysosomal enzyme which hydrolyzes cerebroside sulphate. Arylsulphatase B (ASB),also a lysosomal enzyme, which hydrolyzes the sulphate ester group from N-acetylgalactosamine 4-sulphate group of dermatan sulphate.
Functional site
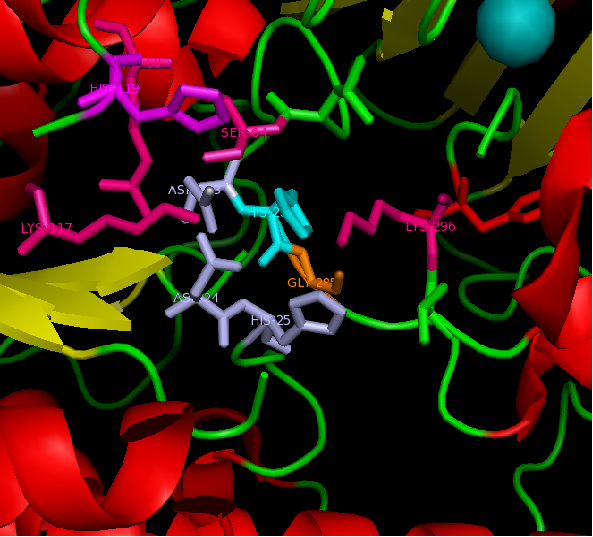
MSA data revealed some conserved residues on the sequence. They were mapped on the three dimentional structure.
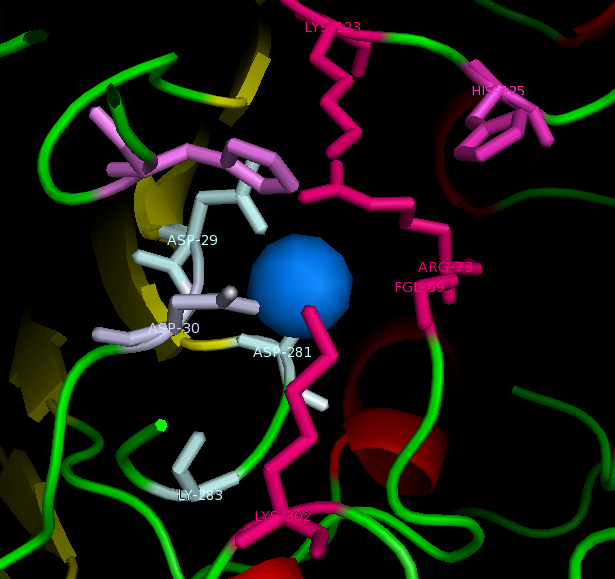
Figure 1: Catalytic site of Arylsulfatase A (ASA) with the Magnesium ion bound. However H229 is only conserved within Arylsulfatase A of different species.
figure 2: Catalytic site of Arylsulfatase K (ASK) with conserved catalytic residues. Residues marked in grey are possible to be involved in divalent metal binding , while residues marked in magenta are highly conserved with those of Arylsulfatase A (ASA) and steroid sulfatase (STS). Histidine (H) shown in cyan is not strictly consetved in MSA, but the only available nucleophile in the close proximity.
Highly conserved regions of the protein revealed in MSA are highlighted in blue, while ligands; Zn and Cl; are shown in green and grey respectively. However, region of high conservation seems to be physically further from ligand binding sites. Maybe suggesting that ligands only play a role in stabilizing the protein conformation, hence the active site; for the enzyme to perform it's catalytic functions.
.
Structural alignment
- Three dimentional structure of arylsulfatase was aligned with other available structures using DALI server (structural alignment). Results are shown in 'figure 4'.
No: Chain Z rmsd lali nres %id Description 1: 3b5q-A 73.6 0.0 464 464 100 MOLECULE: PUTATIVE SULFATASE YIDJ; 2: 3b5q-B 70.2 0.3 464 467 100 MOLECULE: PUTATIVE SULFATASE YIDJ; 3: 2qzu-A 35.1 2.5 375 465 25 MOLECULE: PUTATIVE SULFATASE YIDJ; 4: 1fsu 28.7 2.8 344 474 22 MOLECULE: N-ACETYLGALACTOSAMINE-4-SULFATASE; 5: 1n2l-A 28.4 3.0 343 483 25 MOLECULE: ARYLSULFATASE A; 6: 1n2k-A 28.3 3.2 344 482 25 MOLECULE: ARYLSULFATASE A; 7: 1e3c-P 28.2 3.2 344 481 26 MOLECULE: ARYLSULFATASE A; 8: 1e33-P 28.2 3.1 344 480 25 MOLECULE: ARYLSULFATASE A; 9: 1e2s-P 28.2 3.1 343 481 25 MOLECULE: ARYLSULFATASE A; 10: 1e1z-P 28.1 3.2 344 481 26 MOLECULE: ARYLSULFATASE A; 11: 1auk 27.9 3.1 343 481 25 MOLECULE: ARYLSULFATASE A;
Figure 4: Structurally related proteins. (No 1 and 2 are two chains of arylsulfatase K).
- The function of highly related proteins were found using ProFunc
- Putative Sulfatase YIDI hydrolyses sulfuric ester bonds of its substrate hence significant in metabolism.
- Arylsulfatase A has both sulfuric ester hydrolase and phosphoric monoester hydrolase activities.
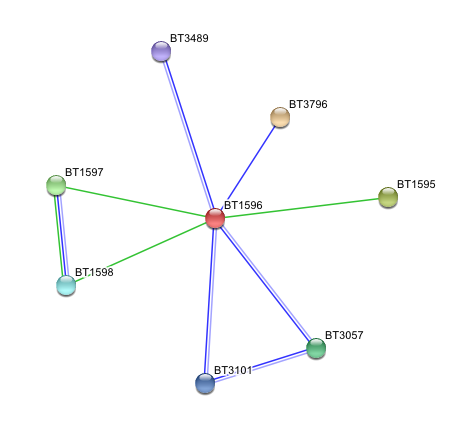
Arylsulfatase K (BT1596) Interactions with other proteins
Input Protein
- BT1596 Putative sulfatase yidJ (481 aa) of Bacteroides thetaiotaomicron.
Predicted Functional Partners
- BT3796: Putative secreted sulfatase ydeN (518 aa).
- BT1595 Transcription termination factor rho (722 aa).
- BT1597 Two-component system sensor histidine kinase (539 aa).
- BT3057 N-acetylgalactosamine-6-sulfatase (508 aa).
- BT1598 Putative two-component system sensor histidine (655 aa).
- BT3101 N-sulphoglucosamine sulphohydrolase (455 aa).
- BT3489 Arylsulfatase B {UniProtKB/TrEMBL-Q8A219} (458 aa).
N-acetylgalactosamine-6-sulfatase cleaves the 6-sulfate groups of N-acetyl-D-galactosamine 6-sulfate units in chondroitin sulfate and D-galactose 6-sulfate units in keratan sulfate.N-sulphoglucosamine sulphohydrolase is also known as heparine sulfamidase, which catalyses the hydrolysis of Sulfur-Nitrogen bonds. N-sulphoglucosamine sulphohydrolase is responsible for the degradation of glucosaminlglycan and glycan structure of extra cellular matrix.
- N-sulfo-D-glucosamine + H(2)O <=> D-glucosamine + sulfate
N-acetylgalactosamine-4- sulfatase (Arylsulfatase B) hydrolyse the sulfate ester group from N-acetylgalactosamine 4-sulfate of dermatine sulfate. Deficiency of ASB cause a rare mucopolysaccharidosis (MPS IV; Maroteaux-Lamy syndrome)
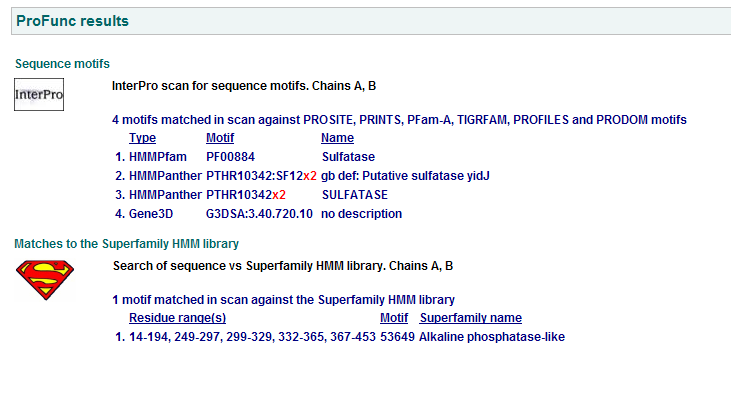
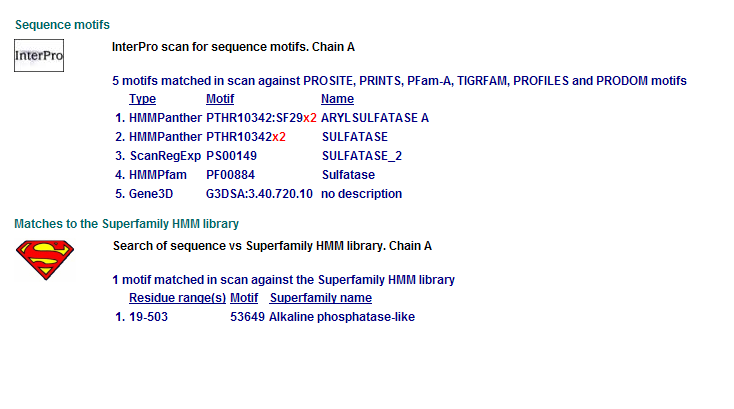
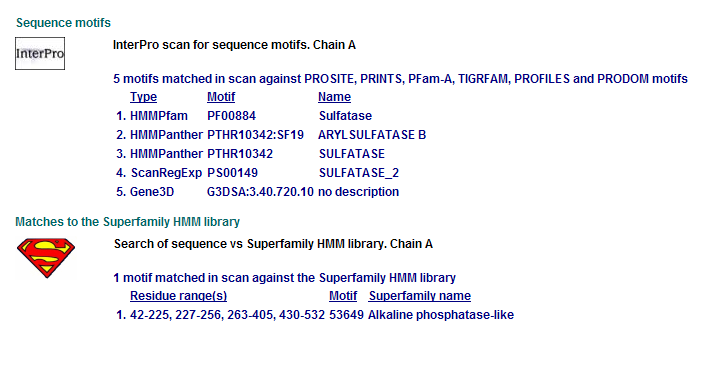
Comparison of Motifs
1 Putative sulfatase yidj motifs
2 Arylsulfatase A. Synonym: cerebroside-3-sulfate-sulfatase motifs
3 N-acetylgalactosamine-4-sulfatase motifs. (Chain: a. Synonym: arylsulfatase b, asb, 4-sulfatase)
Motifs PF00884 and PTHR10342 are conserved among all three proteins. This is indicative of a some structural similarity in their substrates. These proteins may carry out a similar catalytic reaction or participate in slightly different aspects of a common catalytic activity. The second possibility seems to be stronger considering all three enzymes are found in tha same metabolic pathway, in close proximity to each other. All the resulted motifs are found in a sulfatase enzyme while they all belong to Alkaline phosphatase-like protein superfamily.
PTHR10342 is responsible for synthesis and breakdown of sulfur containing compounds, and sulfur-dependent redox reactions.
Click here to go Back