Results 2ece: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[ | == STRUCTURAL ANALYSIS == | ||
'''SBP 1 Amino Acid FASTA FORMAT Sequence''' | |||
>2ECE:A|PDBID|CHAIN|SEQUENCE | |||
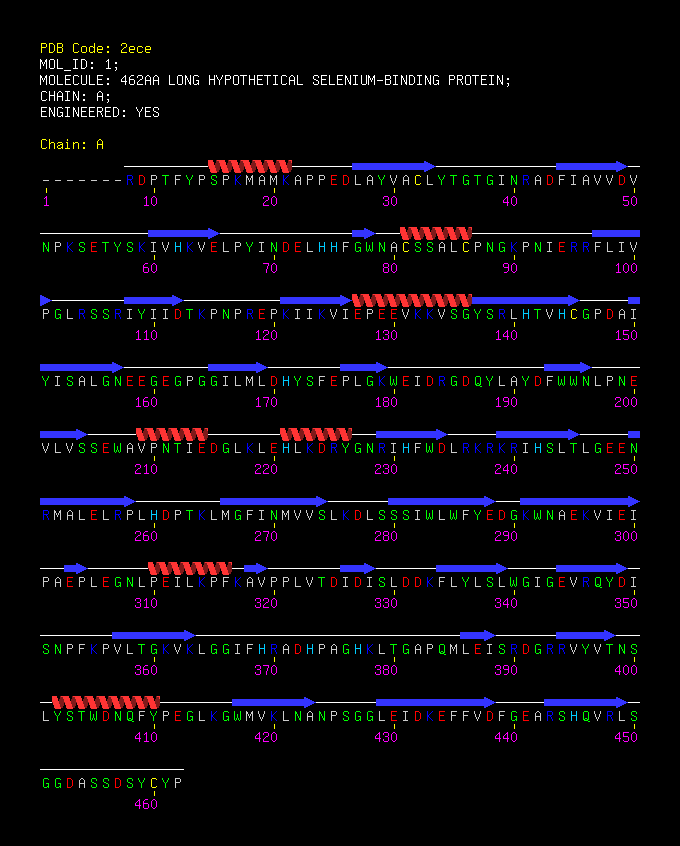
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKIVHKVELPYINDELHHFGWNA | |||
CSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREPKIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEG | |||
EGPGGILMLDHYSFEPLGKWEIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR | |||
IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEIPAEPLEGNLPEILKPFKAVP | |||
PLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTGKVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNS | |||
LYSTWDNQFYPEGLKGWMVKLNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP | |||
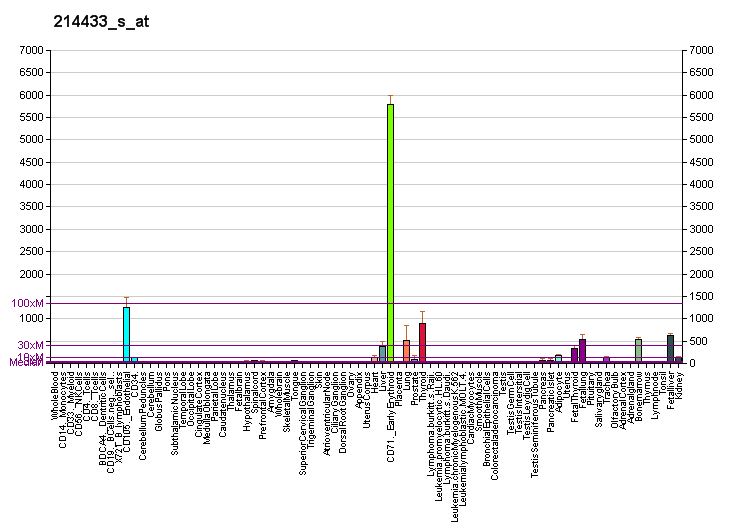
[[Image:PBB_GE_SELENBP1_214433_s_at_fs.png|frame|border|center|Gene expression of the SELENBP1 gene based on gene atlas of the mouse and human protein -encoding transcriptomes-proc. Natl. Ac. Sci. 101 (16) 6062-7]] | |||
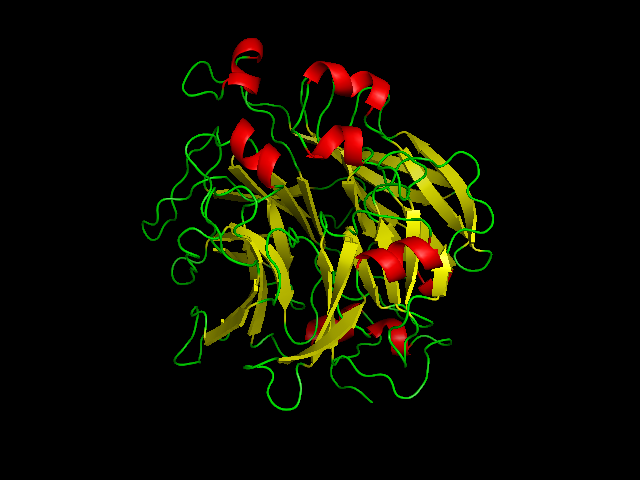
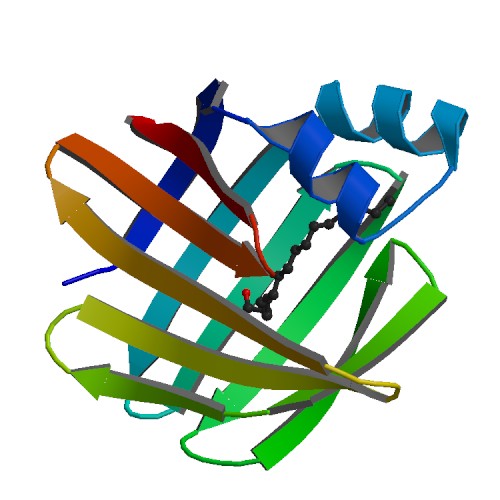
[[Image:2ece_bio_r_500structure.jpg|frame|border|left|'''Figure 1''' X-ray structure of hypothetical selenium-binding protein from ''Sulfolobus tokodaii'', ST0059 ( http://www.proteopedia.org/wiki/index.php/2ece ) and the JenaLib Jmol viewer showing SBP 1 secondary structure spinning [http://www.imb-jena.de/cgi-bin/3d_mapping.pl?CODE=2ece&MODE=asymmetric]] | |||
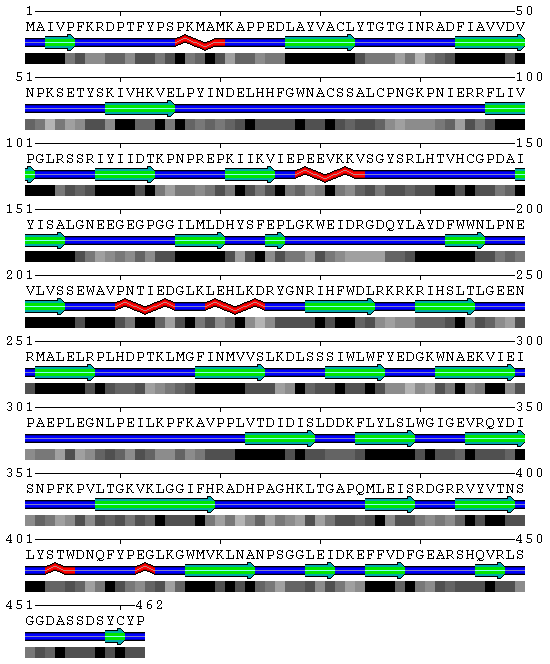
[[Image:SABLE.gif|frame|border|left|'''Figure 2''' SBP amino acid structure prediction derived from the SABLE server prediction- '''Query name''': SBP 1 ( 2ECE ) | |||
Structure prediction by SABLE | |||
'''Data source''': Derived from the SABLE server prediction | |||
]] | |||
'''WARNING!''' Given sequence appeared to be a soluble protein, no TM domains found! | |||
Output format is the following: | |||
'''1st line''' -> residue numeration | |||
'''2nd line''' -> query amino acid sequence | |||
'''3rd line''' -> trans-membrane domain prediction (T-TM region, '''N'''-soluble part) | |||
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKI | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
VHKVELPYINDELHHFGWNACSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREP | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
KIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEGEGPGGILMLDHYSFEPLGKW | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
EIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEI | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
PAEPLEGNLPEILKPFKAVPPLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTG | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
KVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNSLYSTWDNQFYPEGLKGWMVK | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
LNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP | |||
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN | |||
'''Figure 3''' SABLE server results. | |||
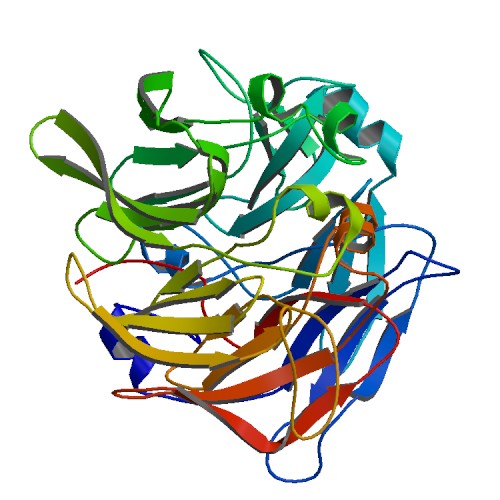
[[Image:movie0001.png|frame|border|left|'''Figure 4''' Shows structural helices(red) and beta sheets (yellow) of SBP designed from pymol | |||
]] | |||
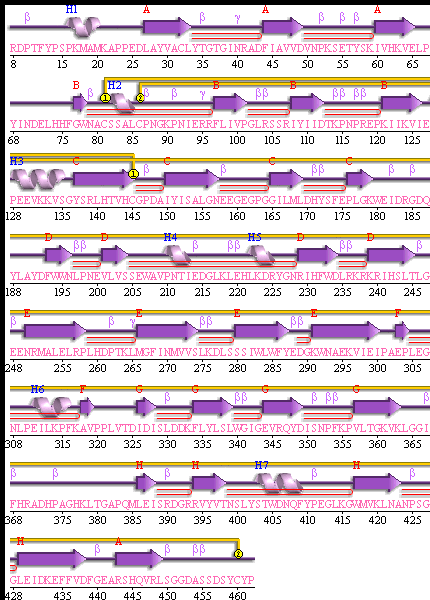
[[Image:secondary structure.gif|frame|border|left|'''Figure 5''' PDBsum of structural representation of SELENBP1 showing disulphide bonds networks (yellow), loops, helices (labelled blue) and beta sheets ( labelled Red)]] | |||
[[Image:pdb_cartoon_2ece.png|frame|border|left|'''Figure 6''' PDBsum shows the beta and a- helices of SELENBP1 ]] | |||
STRUCTURAL COMPARISONS | |||
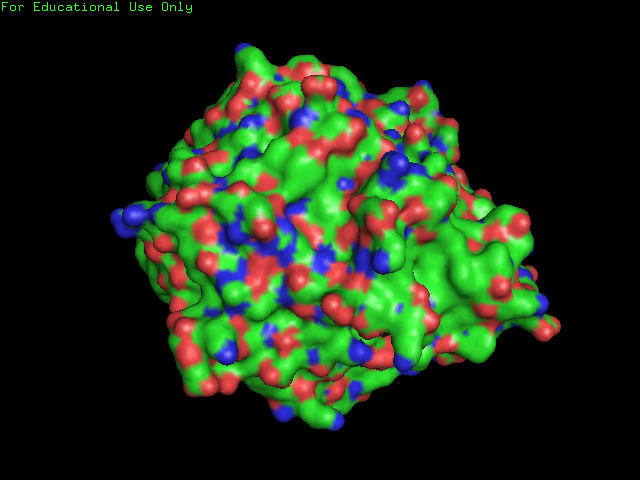
[[Image:surface of 2ece.png|frame|border|left|'''Figure 7''' Surface Structure of SBP: Note the few clefts compared to the DNA isomerase shown below.]] | |||
[[Image:1YUA SURFACE STRUCTURE.JPG|frame|border|left|'''Figure 8''' pdb (1yua)-Topo-Zn DNA isomerase surface structure[http://www.ncbi.nlm.nih.gov/entrez/query/static/gifs/entrez_struc.gif] ]] | |||
[[Image:Rat fatty acid binding protein 2IFB.jpg|frame|border|left|'''Figure 9''' Rat fatty acid binding protein 2IFB, a was found to have 92.5% homology to 14 KDa Selenium binding protein purified from rat liver using column chromatography and SDS-Gel techniques.([[http://compbio.chemistry.uq.edu.au/mediawiki/index.php/References_2ece ref 3]])]] | |||
Explore SBP features and structural summary here [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=61601].The domains of SBP are shown here [http://www.ncbi.nlm.nih.gov/sites/entrez?db=domains&cmd=search&term=2ece] | |||
Notice how the domains are similar to the putative Isomerase domains of E.coli on '''Figure 10'''below. | |||
'''1RI6 DOMAINS''' | |||
[[Image:1RI6 A 0.png|frame|800x800px|left|full structure]] | |||
[[Image:1RI6 A 1.png|frame|left|Domain 1]] | |||
[[Image:1RI6 A 2.png|frame|left|Domain 2]] | |||
[[Image:1RI6 A 3.png|frame|left|Domain 3]] | |||
'''2ECE DOMAINS''' | |||
[[Image:2ece A 3.png|frame|left|Domain 3]] | |||
[[Image:2ece A 2.png|frame|left|Domain 2]] | |||
[[Image:2ece A 1.png|frame|left|Domain 1]] | |||
[[Image:2ece A 0.png|frame|left|full structure]] | |||
[[Image:Ligand of bovine.png|frame|left|'''Figure 11'''Complex Of Bovine Odorant Binding Protein (Obp) With A Selenium Containing Odorant)"Image:Ligand of bovine.png" [[http://compbio.chemistry.uq.edu.au/mediawiki/upload/2/23/Ligand_of_bovine.png]]]] | |||
[[FUNCTIONAL analysis of 2ece]] | |||
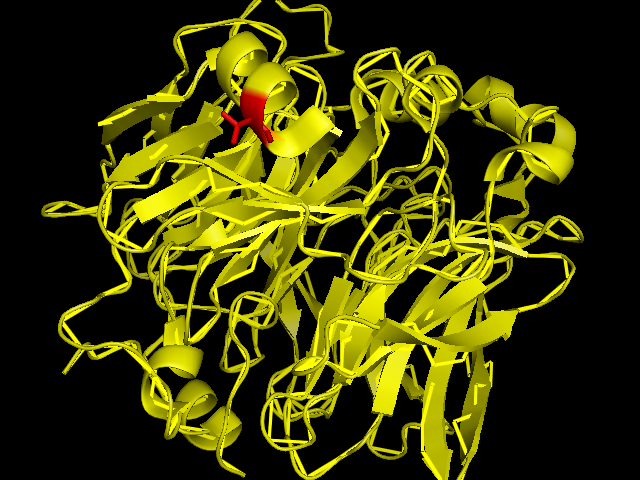
[[Image:SBP with Sele atom.png]] | [[Image:SBP with Sele atom.png]] | ||
The red coloured region is the region carring Se, and it is evident that the Se is incorporated in an amino acid of the helix chain.(Pymol Image) | The red coloured region is the region carring Se, and it is evident that the Se is incorporated in an amino acid of the helix chain.(Pymol Image) | ||
Revision as of 11:00, 9 June 2008
STRUCTURAL ANALYSIS
SBP 1 Amino Acid FASTA FORMAT Sequence
>2ECE:A|PDBID|CHAIN|SEQUENCE
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKIVHKVELPYINDELHHFGWNA CSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREPKIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEG EGPGGILMLDHYSFEPLGKWEIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEIPAEPLEGNLPEILKPFKAVP PLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTGKVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNS LYSTWDNQFYPEGLKGWMVKLNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP
WARNING! Given sequence appeared to be a soluble protein, no TM domains found!
Output format is the following:
1st line -> residue numeration
2nd line -> query amino acid sequence
3rd line -> trans-membrane domain prediction (T-TM region, N-soluble part)
MAIVPFKRDPTFYPSPKMAMKAPPEDLAYVACLYTGTGINRADFIAVVDVNPKSETYSKI
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
VHKVELPYINDELHHFGWNACSSALCPNGKPNIERRFLIVPGLRSSRIYIIDTKPNPREP
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
KIIKVIEPEEVKKVSGYSRLHTVHCGPDAIYISALGNEEGEGPGGILMLDHYSFEPLGKW
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
EIDRGDQYLAYDFWWNLPNEVLVSSEWAVPNTIEDGLKLEHLKDRYGNRIHFWDLRKRKR
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
IHSLTLGEENRMALELRPLHDPTKLMGFINMVVSLKDLSSSIWLWFYEDGKWNAEKVIEI
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
PAEPLEGNLPEILKPFKAVPPLVTDIDISLDDKFLYLSLWGIGEVRQYDISNPFKPVLTG
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
KVKLGGIFHRADHPAGHKLTGAPQMLEISRDGRRVYVTNSLYSTWDNQFYPEGLKGWMVK
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
LNANPSGGLEIDKEFFVDFGEARSHQVRLSGGDASSDSYCYP
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
Figure 3 SABLE server results.
STRUCTURAL COMPARISONS
Explore SBP features and structural summary here [3].The domains of SBP are shown here [4]
Notice how the domains are similar to the putative Isomerase domains of E.coli on Figure 10below.
1RI6 DOMAINS
2ECE DOMAINS
The red coloured region is the region carring Se, and it is evident that the Se is incorporated in an amino acid of the helix chain.(Pymol Image)