Results (1zkd)
Structure of Hypothetical Protein LOC55471 Isoform 1
Dali shows a few proteins with similar structure to 1zkd. They are 2ex4 and 1im8 which shows the highest Z-value of 11.7 and 11.6 respectively. 2ex4 is a human methyltransferase with S-adenosylhomocysteine and 1im8 is found to be a methyltransferase with a bound S-adenosylhomocysteine from the crystal structure of YecO from Haemophilus influenzae (HI0319).
SUMMARY: PDB/chain identifiers and structural alignment statistics
NR. STRID1 STRID2 Z RMSD LALI LSEQ2 %IDE REVERS PERMUT NFRAG TOPO PROTEIN
1: 3027-A 1zkd-A 56.8 0.0 349 349 100 0 0 1 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION duf185 (rhodops 2: 3027-A 2ex4-A 11.7 3.0 185 221 12 0 0 22 S TRANSFERASE adrenal gland protein ad-003 (homo sapien 3: 3027-A 1im8-A 11.6 3.2 178 225 10 0 0 18 S TRANSFERASE yeco (methyltransferase, hypothetical pro 4: 3027-A 2gb4-A 10.8 3.3 184 231 13 0 0 19 S TRANSFERASE thiopurine s-methyltransferase (thiopurine 5: 3027-A 2fk7-A 10.7 3.8 186 277 14 0 0 19 S TRANSFERASE methoxy mycolic acid synthase 4 (mycobact 6: 3027-A 2ob1-A 10.2 3.9 196 319 9 0 0 23 S TRANSFERASE leucine carboxyl methyltransferase 1 (prot 7: 3027-A 2f8l-A 10.0 3.7 182 318 8 0 0 22 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION hypothetical pro 8: 3027-A 2avn-A 10.0 3.7 183 242 11 0 0 20 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION ubiquinoneMENAQU 9: 3027-A 2bzg-A 9.9 3.4 182 226 13 0 0 20 S TRANSFERASE thiopurine s-methyltransferase (thiopurine 10: 3027-A 2aot-A 9.8 4.3 182 285 13 0 0 23 S TRANSFERASE histamine n-methyltransferase (hmt) (homo
1zkd is an unknown protein, and by using proteins similar to it, the functions of this unknown protein can be predicted. With 2ex4 and 1im8 showed by Dali to be the most similar, other tools are used to determine the similarity. Combinatorial Extension Method is used. Below shows the sequence alignment and structure alignment of the unknown protein with the proteins obtained from dali:
Alignment with 2ex4
1ZKD:A 24/25 WRYXELCLGHPEHGYYV--TRDPLGREGDFTTSPEISQXFGELLGLWSASVWKAAD-EPQ 2EX4:A 24/7 IEDEKQFYS----KAKTYWKQIPPTVDGMLGGYGHISSIDINSSRKFLQRFLREGPNKTG 1ZKD:A 81/82 TLRLIEIGPGRGTXXADALRALRVLPILYQSLSVHLVEINPVLRQKQQTLLAGI-RNIHW 2EX4:A 80/64 TSCALDCGAGIGRITKRLLLPL--------FREVDMVDITEDFLVQAKTYLGEEGKRVRN 1ZKD:A 140/141 HD-----SFEDVPEGPAVILANEYFDVLPIHQAIKRETGWHERVIEIGASGELVFGVAAD 2EX4:A 132/116 YFCCGLQDFTPEPDSYDVIWIQWVIGHLT------------------------------- 1ZKD:A 195/196 PIPGFEALLPPLARLSPPGAVFEWRP--DTEILKIASRVRDQGGAALIIDYG--HLRSDV 2EX4:A 161/145 ------------------------DQHLAEFLRRCKGSL-RPNGIIVIKDNMAQE----- 1ZKD:A 251/252 GDTFQAIASHSYADPLQHPGRADLTAHV---DFDALGRAAESIGARAHGPVTQG 2EX4:A 191/175 GVILDD---------------VDSSVCRDLDVVRRIICSAG---LSLLAEERQE
Alignment with 1im8 1ZKD:A 59/60 QXFGELLGLWSASVWKAADEPQTLRLIEIGPGRGTXXADALRALRVLPILYQSLSVHLVE 1IM8:A 42/40 SNIITAIGXLAERFV-----TADSNVYDLGCSRGAATLSARRNI-----NQPNVKIIGID 1ZKD:A 119/120 INPVLRQKQQTLLAGI---RNIHWHD--SFEDVPEGPAVILANEYFDVLPIHQAIKRETG 1IM8:A 92/90 NSQPXVERCRQHIAAYHSEIPVEILCNDIRHVEIKNASXVILNFTLQFLP---------- 1ZKD:A 174/175 WHERVIEIGASGELVFGVAADPIPGFEALLPPLARLSPPGAVFEWRP--DTEILKIASRV 1IM8:A 142/140 ---------------------------------------------PEDRIALLTKIYEGL 1ZKD:A 232/233 RDQG--GAALIIDYG 1IM8:A 157/155 ---NPNGVLVLSEKF
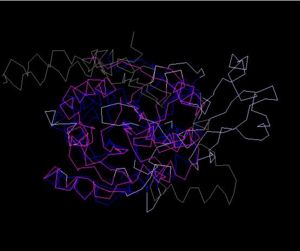
Structure Alignment with 2ex4 (Blue:1zkd, Purple: 2ex4)
Alignment Length: 294
Gaps (average per molecule): 53.5
Sequence Identity: 14.4%
RMSD min – max: 3.03A
Return to Report on 1zkd