Paper
Evolution, Structure and Function of N-acetylneuraminic Acid Phosphatase
Jason Cheong Wen Leong (s41235935), Yau Heen wai (s41286272), Lim Junxian (s41313011)
Abstract
N-acetylneuraminic acid phosphatase a novel protein investigated by our group. With its structure and sequence known, the function was
assumed to be a part of the enormous family of haloacid dehalogenase-like hydrolases. It represent the family of predicted small molecule
phosphatases related by sequence cleave sites and reactions in the genomes of bacteria, archaea, and eukaryotes. Many have evolved to be used
for specific biological functions within individual organism
Introduction
The novel protein investigated by our group is N-acetylneuraminic acid (Neu5Ac) phosphatase, it was first release on Protein Data Bank
(PDB) on 18th April 2006, named 2gfh. Mus muscular (mouse) was used as the source of the gene and Escherichia coli was the
vector used to express the novel protein. In Homo sapiens (man), it was known to be as N-acetylneuraminate 9-phosphate (Neu5Ac-9-P)
phosphatase haloacid dehalogenase (HAD)-like hydrolase domain containing protein 4. Other aliases of the novel protein include C20orf147, NANP
and HDHD4. The gene encoding the protein was found to be on chromosome 20; location 20p11.1.
Neu5Ac-9-P phosphatase belongs to a large family of haloacid dehalogenase (HAD)-like hydrolases. The enzymes found within this classification
possess varied types of cleavage activities. Although many of its members are related by sequence cleave sites and reactions, many have evolved
to be used for specific biological functions within individual organisms.
These small molecule phosphatase enzymes have been found to exists in the various domains of life — Bacteria, Archaea, and Eucarya. The number
of genes found within each organism is varied from bacteria to eukaryotes. Bacterial Neu5Ac synthase and mammalian Neu5Ac-9-P synthase are
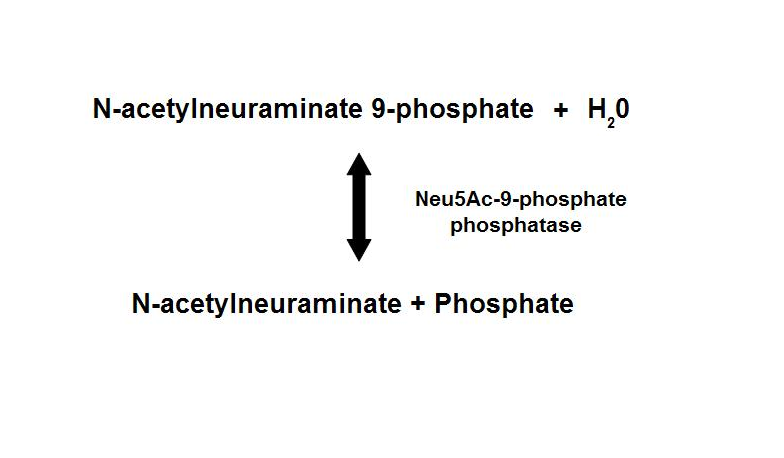
homologous proteins, sharing about 35% sequence identity1. Neu5Ac-9-P phosphatase dephosphorylates Neu5Ac-9-P to form Neu5Ac, the
main form of sialic acid.
Figure 1. Dephosphorylation of Neu5Ac-9-P is a reversible reaction with an end product of Neu5Ac (sialic acid) and a free phosphate.
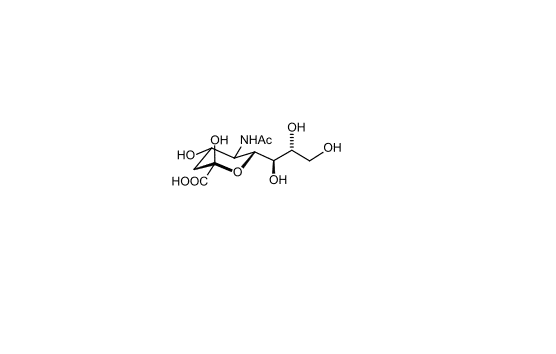
Sialic acids are nine-carbon sugars with a carboxylate group that are found as components of many glycoproteins, glycolipids, and
polysaccharides in animals, viruses, and bacteria. The main form of sialic acid, Neu5Ac, is often present as the terminal sugar of N-
glycans on glycoproteins and glycolipids and plays an important role in protein–protein and cell–cell recognition 2; 3.
Figure 2. Chemical structure of sialic acid.(http://en.wikipedia.org/wiki/Sialic_acid)
Sialic acids are found widely distributed in animal tissues and in bacteria, especially in glycoproteins and gangliosides. The amino group
bears either an acetyl or a glycolyl group. Sialic acid consists of acetylated, sulfated, methylated, and lactylated derivatives and is a large
family of more than 50 members 4.
Results
Query Sequence
The amino acid query sequence of 2gfh protein (Figure 3) from Mus musculus is obtained from Genbank.
1 mgsdkihhhh hhmglsrvra vffdldntli dtagasrrgm levikllqsk yhykeeaeii
61 cdkvqvklsk ecfhpystci tdvrtshwee aiqetkggad nrklaeecyf lwkstrlqhm
121 iladdvkaml telrkevrll lltngdrqtq rekieacacq syfdaivigg eqkeekpaps
181 ifyhccdllg vqpgdcvmvg dtletdiqgg lnaglkatvw inksgrvplt sspmphymvs
241 svlelpallq sidckvsmsv
Figure 3. The 260 amino acid sequence of 2gfh protein.
Sequence Homology
From the BlastP similarity was used for comparison as these had shown higher homology to the query sequence sequence search, a total of 500 proteins were yielded.
Only a total of 38 proteins, in contrast with the remainder of the search results.
These proteins were chosen according to their bit scores and E-values. Two more outlier partial sequences contributing to poor overall alignment (huge deletion gaps) were subsequently removed. The remaining 36 sequences were used for the generation of the phylogenetic tree (and bootstrapped tree as well).
Multiple Sequence Alignment
The following multiple sequence alignment (MSA) was obtained (Figure 4).
From the alignments, gi|10888xy and gi|10888yz are representative of gi|108881764 and gi|108881765 respectively. Both these hypothetical proteins belong to the mosquito Aedes aegypti.
The identifier numbers for these two proteins were initially changed to an alpha-numeric one, due to the inability of Phylip to generate a tree from the original identifiers. This was due to the fact that the programme only took the first five numeric digits (10888), thereby resulting in a programme error prompt which listed both proteins as duplicates (from the identifier numbers). Both these identifiers were subsequently renamed for the final phylogenetic tree.