Arylformamidase Results
Contents
Structure
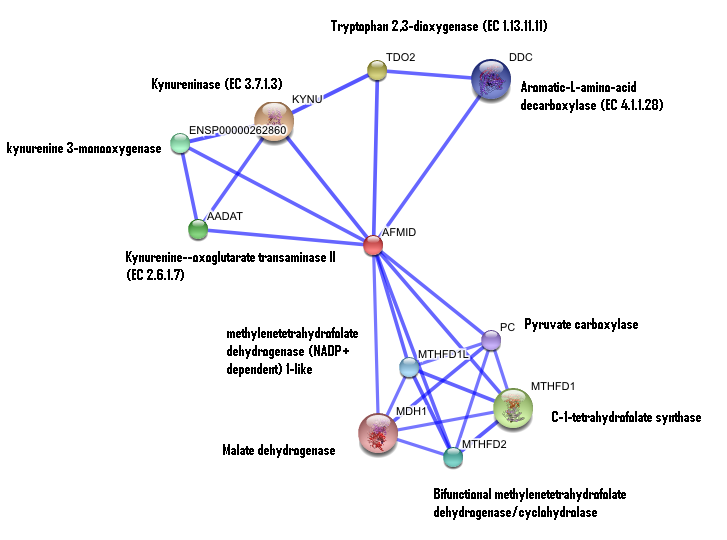
The interaction between the proteins have been determined from curated STRING database (significant score). However there is no significant evidence for:
1- Neighborhood in the genome
2- Gene fusions
3- Cooccurence across genomes
4- Co-Expression
5- Experimental/Biochemical data
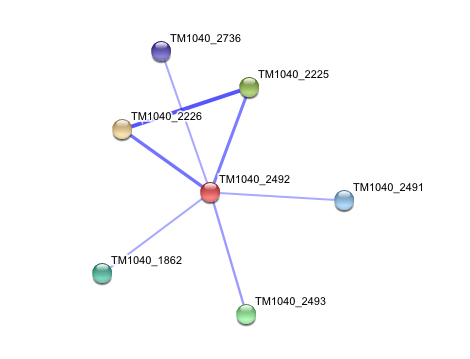
TM1040_2226 Tryptophan 2,3-dioxygenase (279 aa)
TM1040_2225 Kynureninase (396 aa)
TM1040_2493 Succinic semialdehyde dehydrogenase (490 aa)
TM1040_1862 Hypothetical protein (212 aa)
TM1040_2491 Creatinase (402 aa)
TM1040_2736 Transketolase, putative (794 aa)
There is no significant evidence for these interactions (score= ~0.5)
The DALI tool produces proteins that are structurally similar to the protein of interest.
The search result showed similarities to mostly carboxylesterases/hydrolases. Hence there is strong evidence that our protein might also be a carboxylesterase.
PDB link title
Note: Chain B not shown
From PDB ProteinWorkshop 1.5
File:Carboxylesterase (archaeon).txt
Both of the above Archaeal carboxylesterases' chains exist as monomers (from literature). Hence it is expected that our protein exists as a monomer but during crystalization it interacts with its chains.
The secondary structure shows the conservation of the order of different conformation types between the protein of interest and the archaeal carboxylesterases.
Images from PDBsum
The above image shows the conserved residues of the catalytic triad in arylformamidase, with the unknown ligand (Blue) protruding from a surface groove. The residues are serine 136, Histidine 241 and Glutamate 214. Note: The actual residue numbers are n+1
Image generated using Pymol
Image generated using Pymol
The catalytic triad in Archaeoglobus fulgidus Carboxylesterase is very close to the ligand which is also present in aryformamidase.
Function
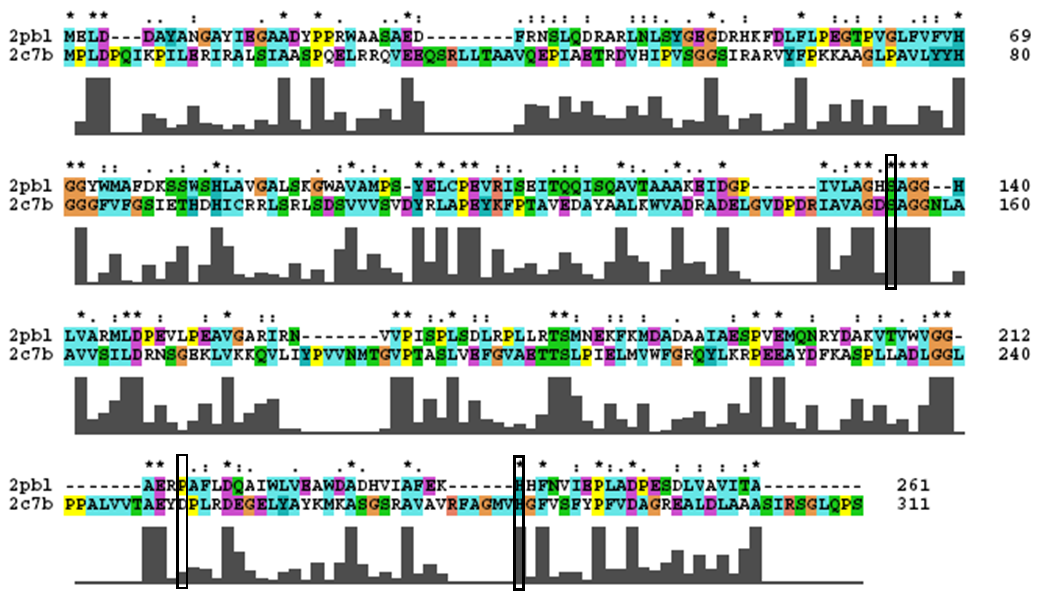
The most similar sequence with functional information available was that of an arylformamidase isolated from the liver of Mus musculus (see figure ...). A functional analysis of this protein has been performed identifying a catalytic triad using site-directed mutagenesis (Pabarcus et al. 2007). Conservation of this catalytic triad with 2pbl was assessed. Both residues S162 and H279 were found to be conserved in relatively conserved regions of the alignment. However, D247 had undergone a semi-conservative substitution. These residues correlated to S136, E214 and H241 of 2pbl which were subsequently located on the tertiary structure and determined to be sufficiently proximal to one another for catalysis (see figure...).
most similar structure - catalytic triad, structure with highlighted
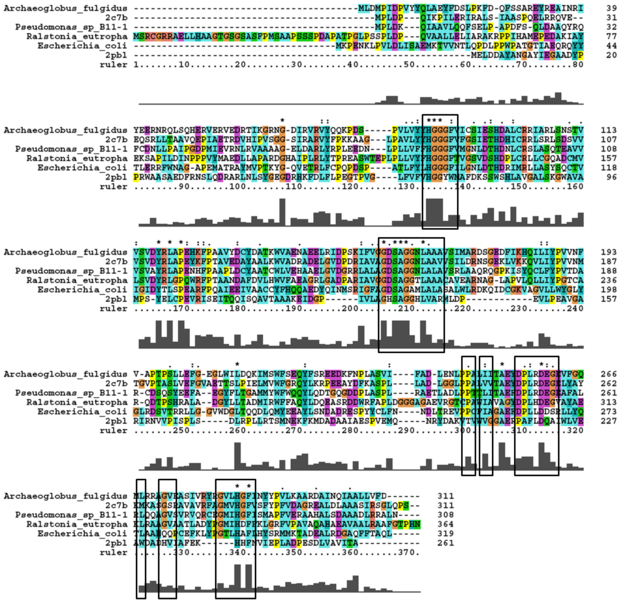
2pbl was found to share most structural similarity with a thermostable carboxylesterase from an uncultured archaeon (PDB ID: 2c7b; see figure ...). 2c7b shares a 16% sequence identity with 2pbl. From its structure, a catalytic triad has been identified (how?). To substantiate any functional similarity between 2pbl and 2c7b, conservation of the 2c7b catalytic triad was analysed (see figure ...). All three residues were found to be conserved, though H... and E... were found to match is less conserved regions.
Sequence & Homology
Figure 1 shows that the query sequence "Arylformamidase" grouped with bacterial sequences, shown cloured in Blue. The bootstrap values reveal low confidence with many of the nodes occurring lower down on the phylogenetic tree revealing a possible explanation for certain closely related species to be grouped into separate clades. However, despite low bootstrap scores, the grouping does reliably separate prokaryotes from eukaryotes and the eukaryotes themsselves are clearly distinguished between yeasts and moulds (shown in Green), plants (Dark Green), invertebrates (Orange) and vertebrates (shown in Red).
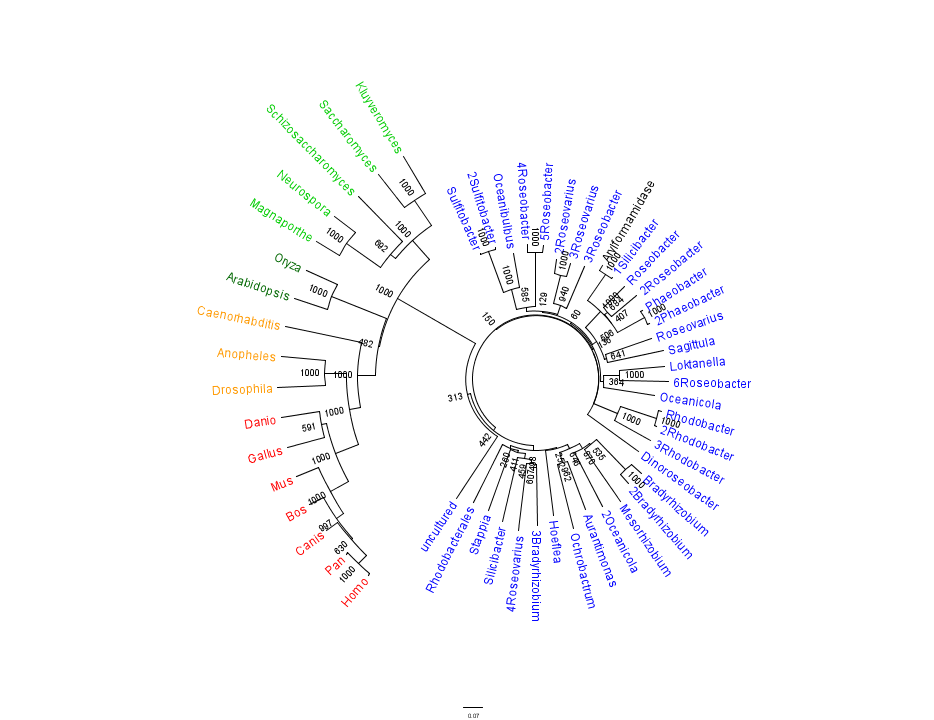
To further elucidate the phylogeny of the Arylformamidase protein, top scoring matches of bacterial homologues were appended with top scoring matches of eukaryotic homologues. Figure 2 is largely consistent with traditional taxonomic groupings of organisms. Specifically, it reveals greater statistical confidence in the separation of prokaryotes (Blue and Green) and eukaryotes (invertebrates are shown in Orange; vertebrates are in Red).
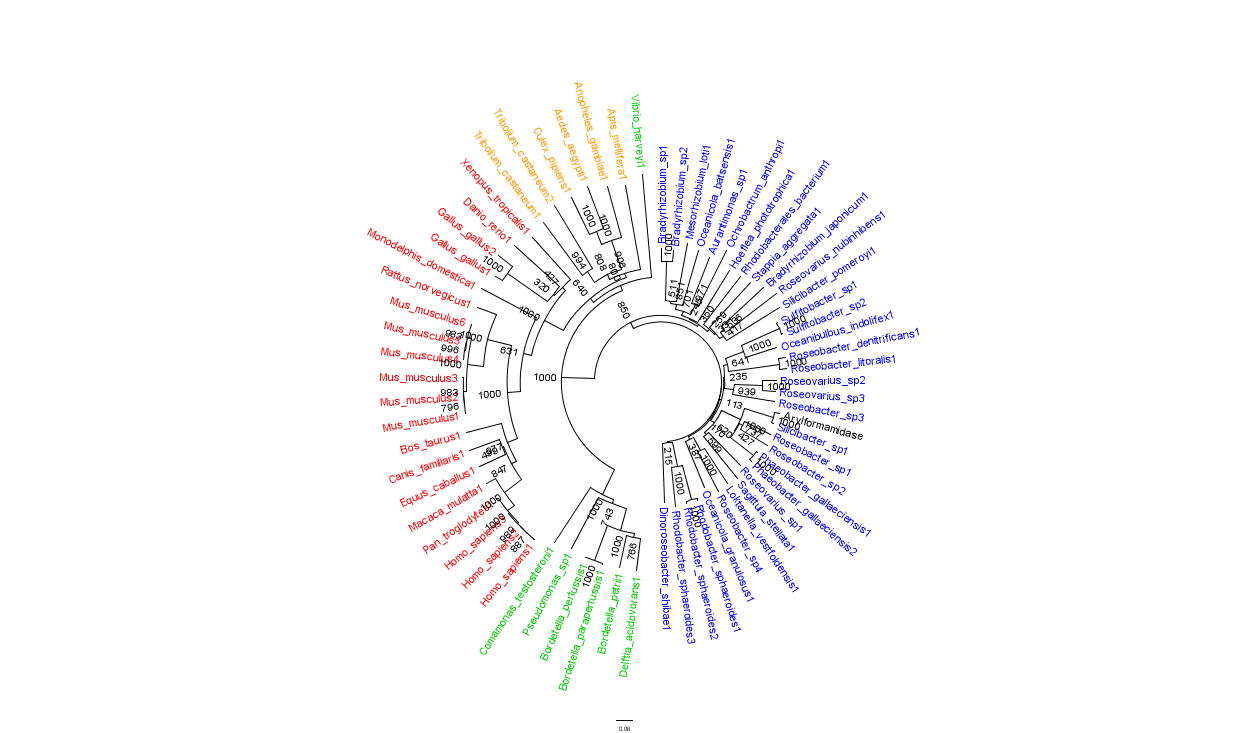
In general, members of the same genus have been grouped together on these phylogenetic trees with some notable exceptions. For instance, Silicibacter, the species from which we derived our protein, occurs on disparate branches of the tree.