Phytanoyl-CoA dioxygenase Sequence and Evolution
From MDWiki
Jump to navigationJump to search
Fasta Sequence
>gi|134105315|pdb|2OPW|A Chain A, Crystal Structure Of Human Phytanoyl-Coa Dioxygenase Phyhd1 (Apo) MACLSPSQLQKFQQDGFLVLEGFLSAEECVAMQQRIGEIVAEMDVPLHCRTEFSTQEEEQLRAQGSTDYF LSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQS MYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSA PGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQP TAELPFPQLYT
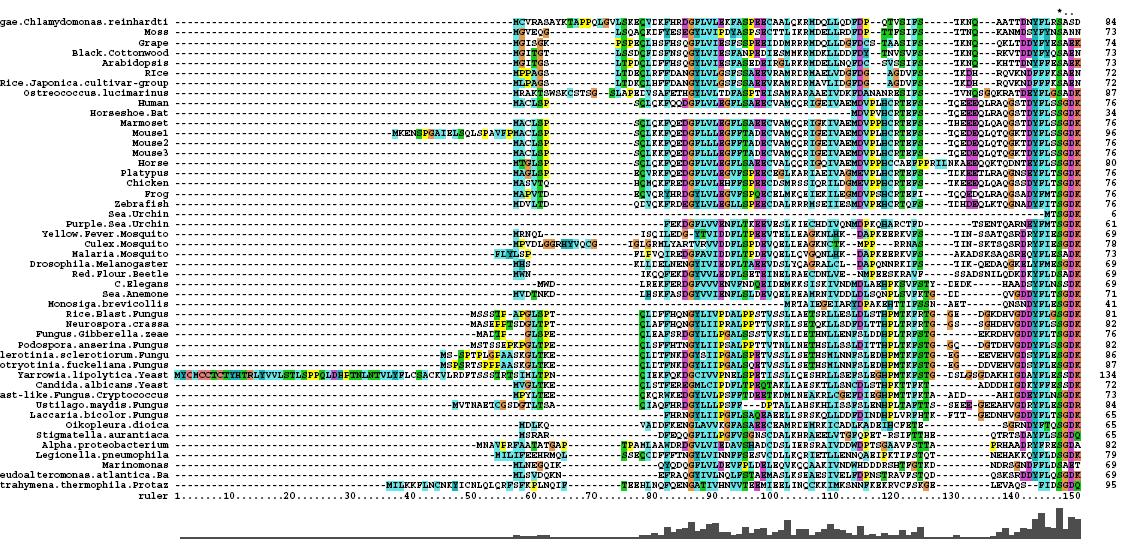
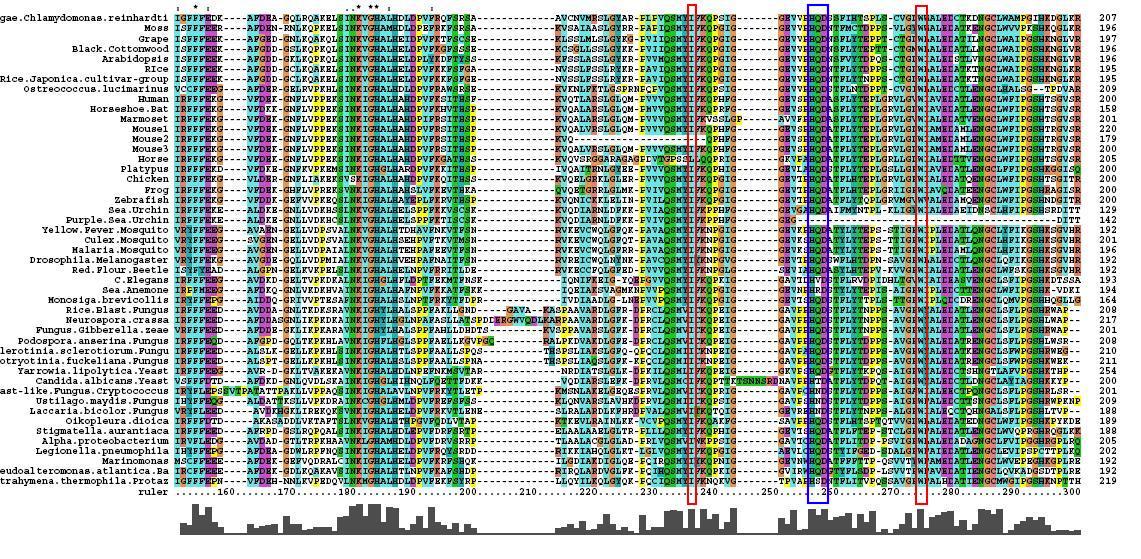
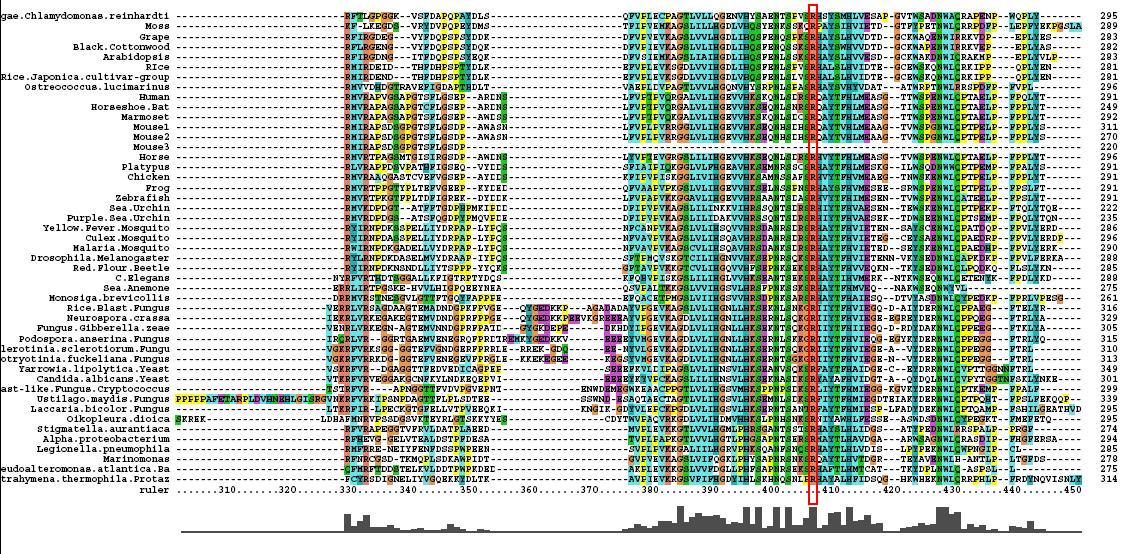
Multiple Sequence Alignment
- 46 analogous sequence compared using ClustalX
- Multiple Sequence Alignment is of high quality and conservation
- Fe2+ and 2-Oxogylterate binding sites shown
Phylogenetic Trees
- Both trees show animals and fungi most highly conserved and plants least conserved
- PhyD required for the breakdown of fatty acids in the diet of animals and fungi
- Original tree similar to consensus tree except for two protozoans
- Monosiga brevicollis and tetrahymena thermophila both have low bootstrap values but some possibility of lateral gene transfer in the case of tetrahymena
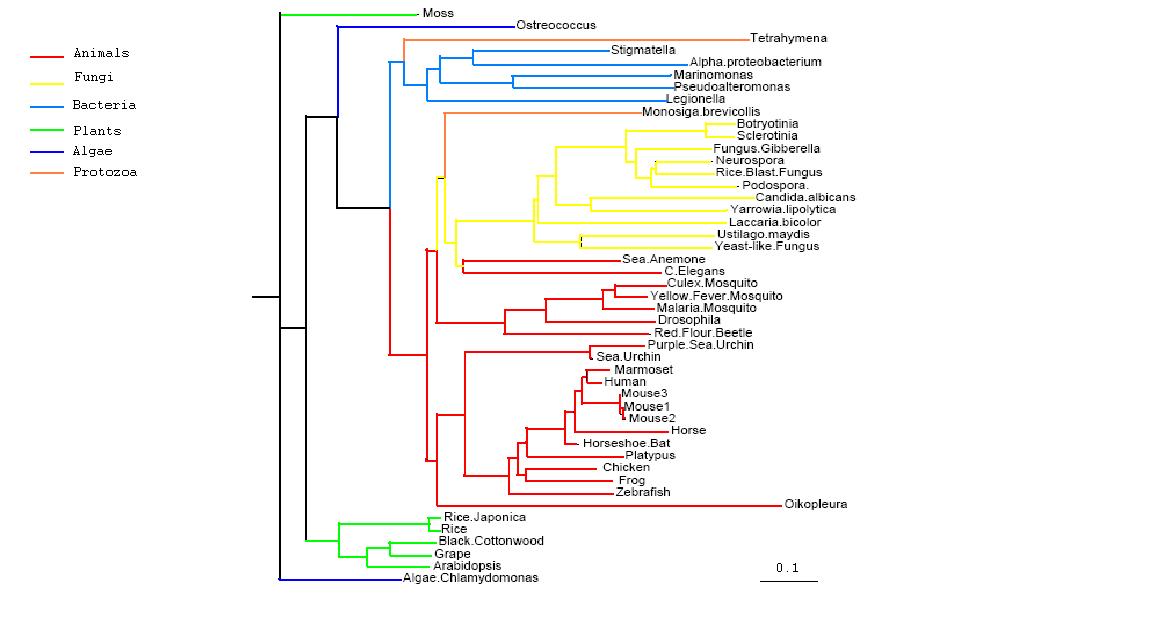
Figure 2.1
Rooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Rooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
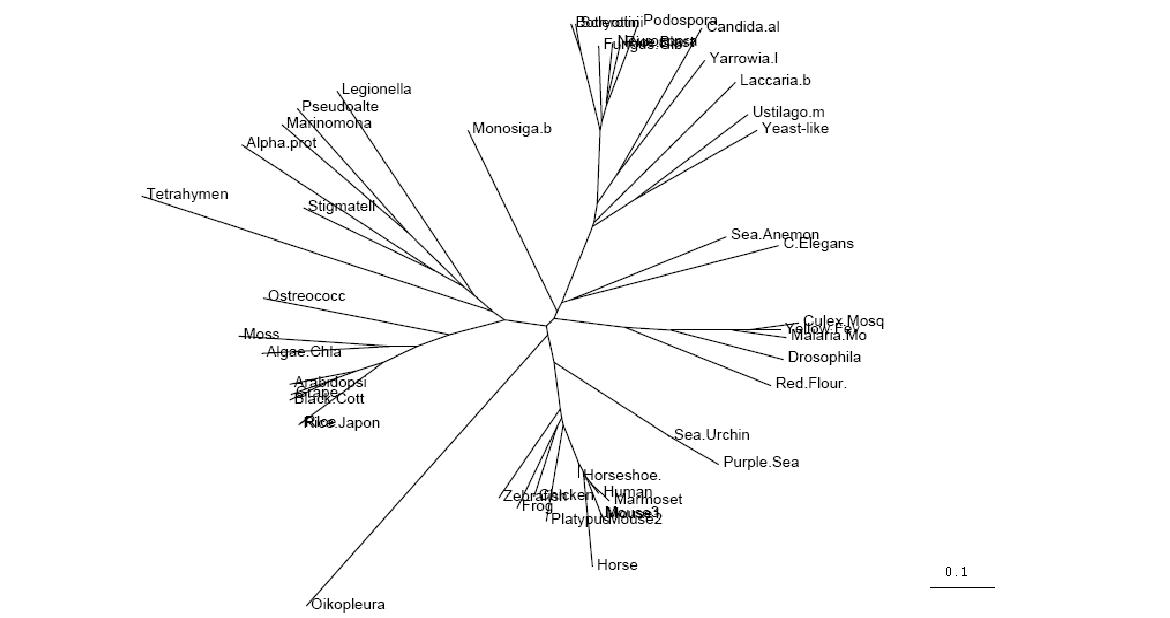
Figure 2.1
Unrooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Unrooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
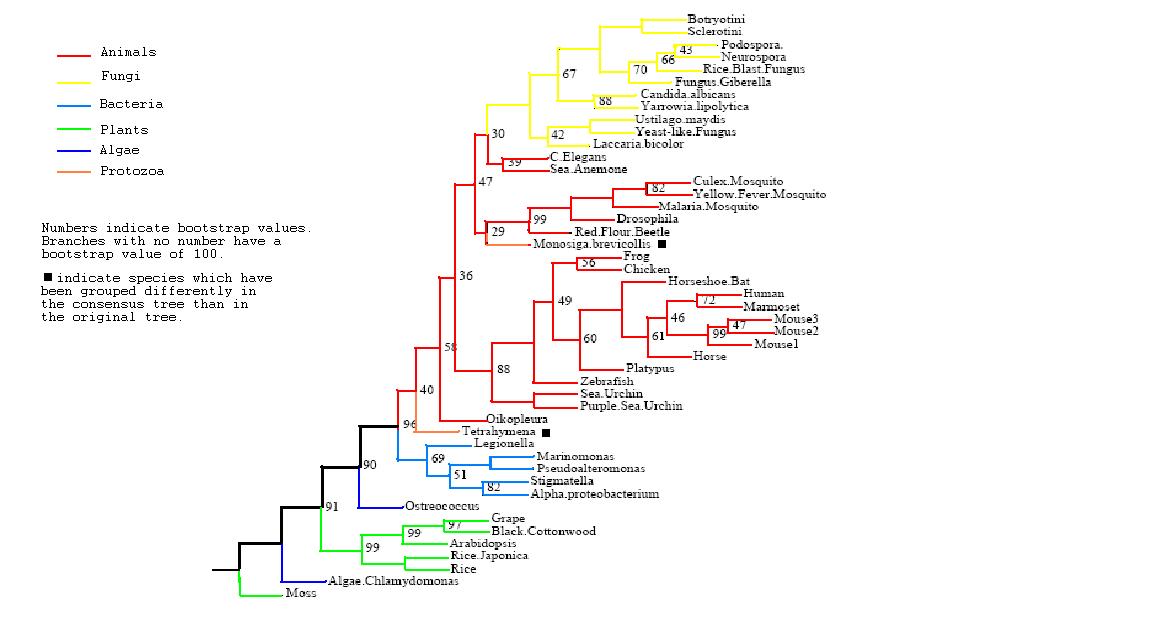
Figure 3
Rooted Consensus Tree with bootstrap values obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Rooted Consensus Tree with bootstrap values obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/