Phytanoyl-CoA dioxygenase Sequence and Evolution
From MDWiki
Jump to navigationJump to search
Fasta Sequence
>gi|134105315|pdb|2OPW|A Chain A, Crystal Structure Of Human Phytanoyl-Coa Dioxygenase Phyhd1 (Apo) MACLSPSQLQKFQQDGFLVLEGFLSAEECVAMQQRIGEIVAEMDVPLHCRTEFSTQEEEQLRAQGSTDYF LSSGDKIRFFFEKGVFDEKGNFLVPPEKSINKIGHALHAHDPVFKSITHSFKVQTLARSLGLQMPVVVQS MYIFKQPHFGGEVSPHQDASFLYTEPLGRVLGVWIAVEDATLENGCLWFIPGSHTSGVSRRMVRAPVGSA PGTSFLGSEPARDNSLFVPTPVQRGALVLIHGEVVHKSKQNLSDRSRQAYTFHLMEASGTTWSPENWLQP TAELPFPQLYT
Multiple Sequence Alignment
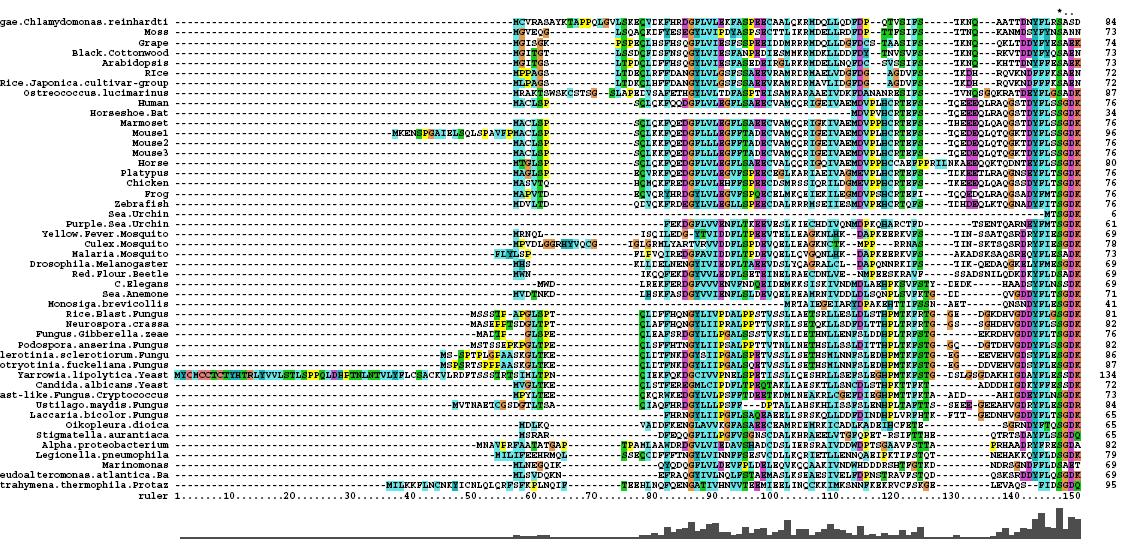
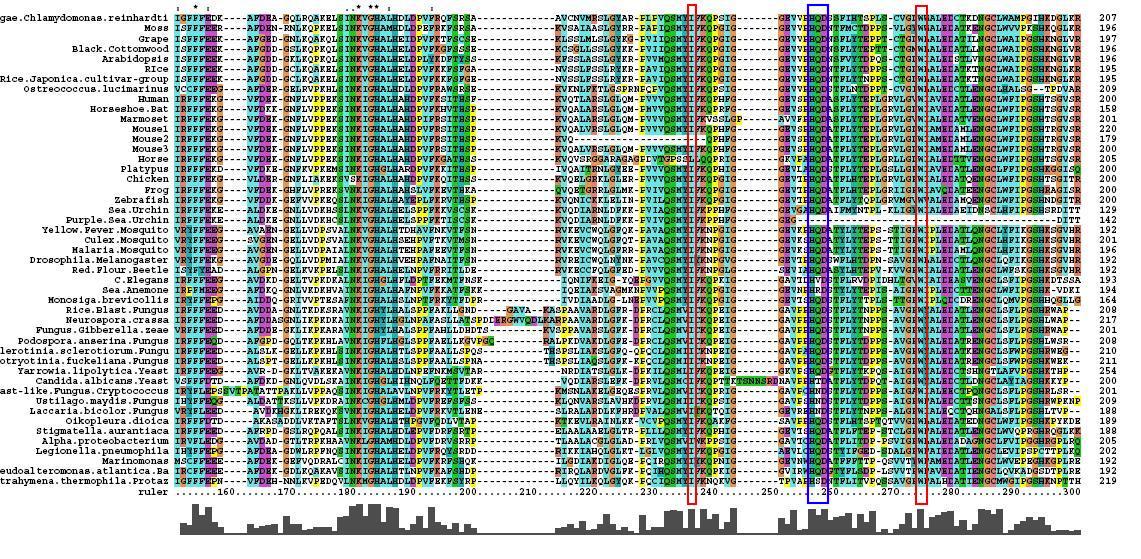
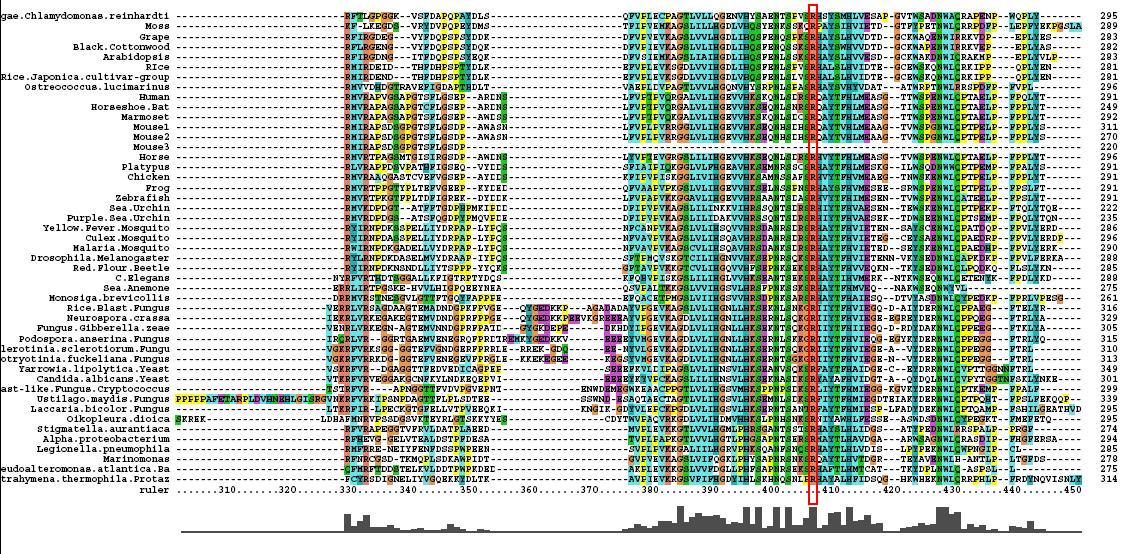
- 46 analogous sequence compared using ClustalX
- Multiple Sequence Alignment is of high quality and conservation
- Fe2+ and 2-Oxogylterate binding sites shown
Phylogenetic Trees
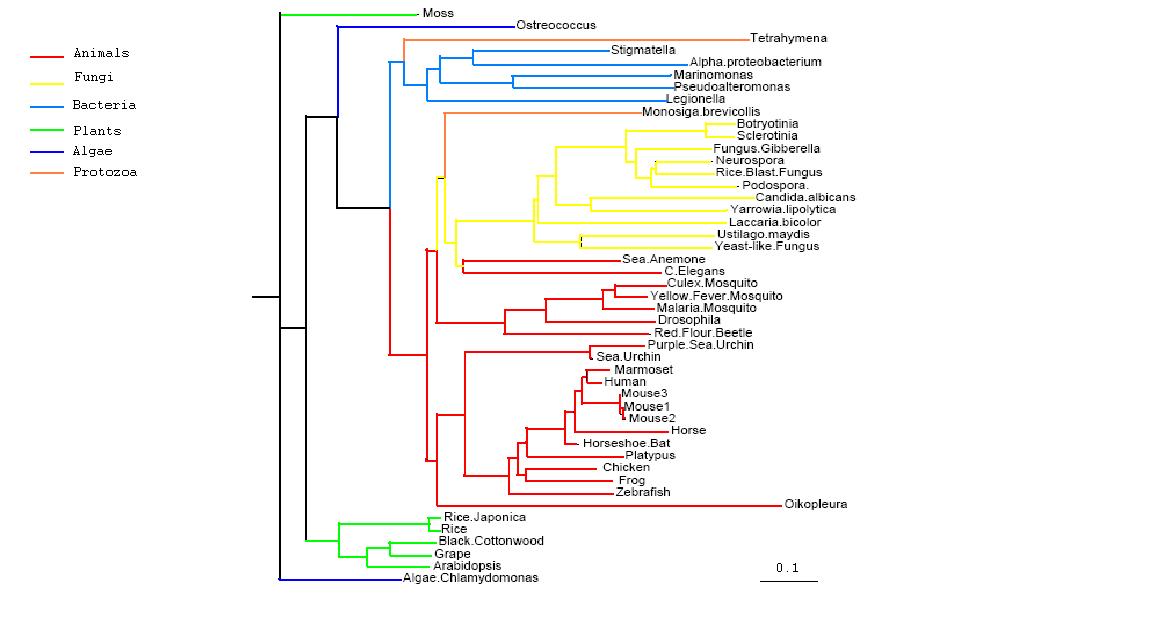
Figure 2.1
Rooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Rooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
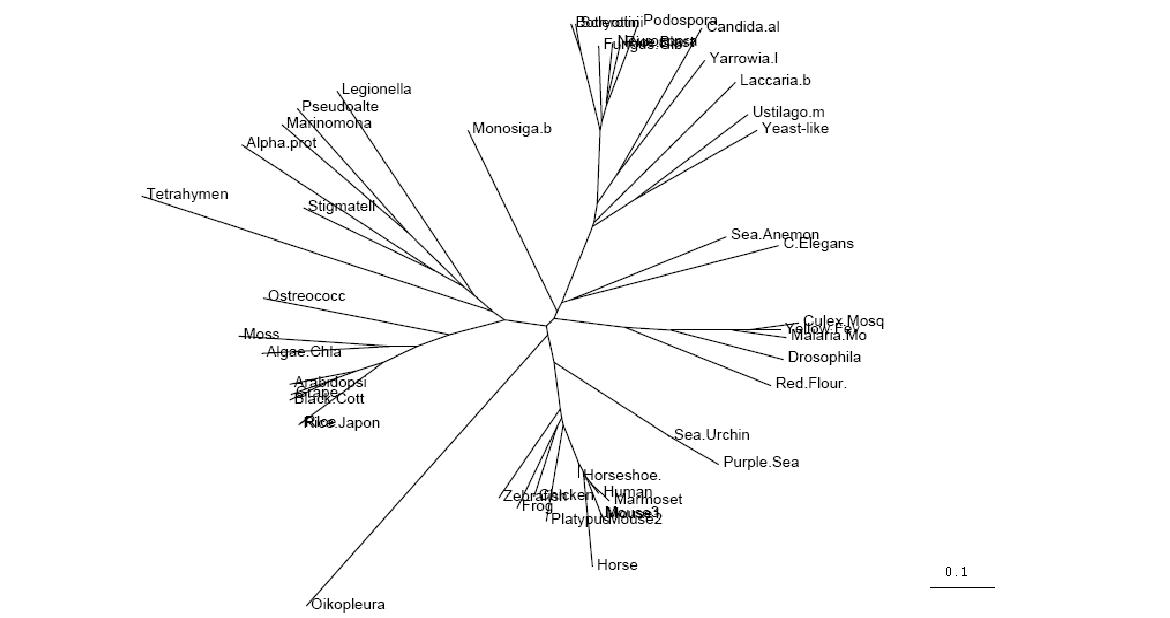
Figure 2.1
Unrooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Unrooted Phylogenetic Tree obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
- Both trees show animals and fungi most highly conserved and plants least conserved
- PhyD required for the breakdown of fatty acids in the diet of animals and fungi
- Original tree similar to consensus tree except for two protozoans
- Monosiga brevicollis and tetrahymena thermophila both have low bootstrap values but some possibility of lateral gene transfer in the case of tetrahymena
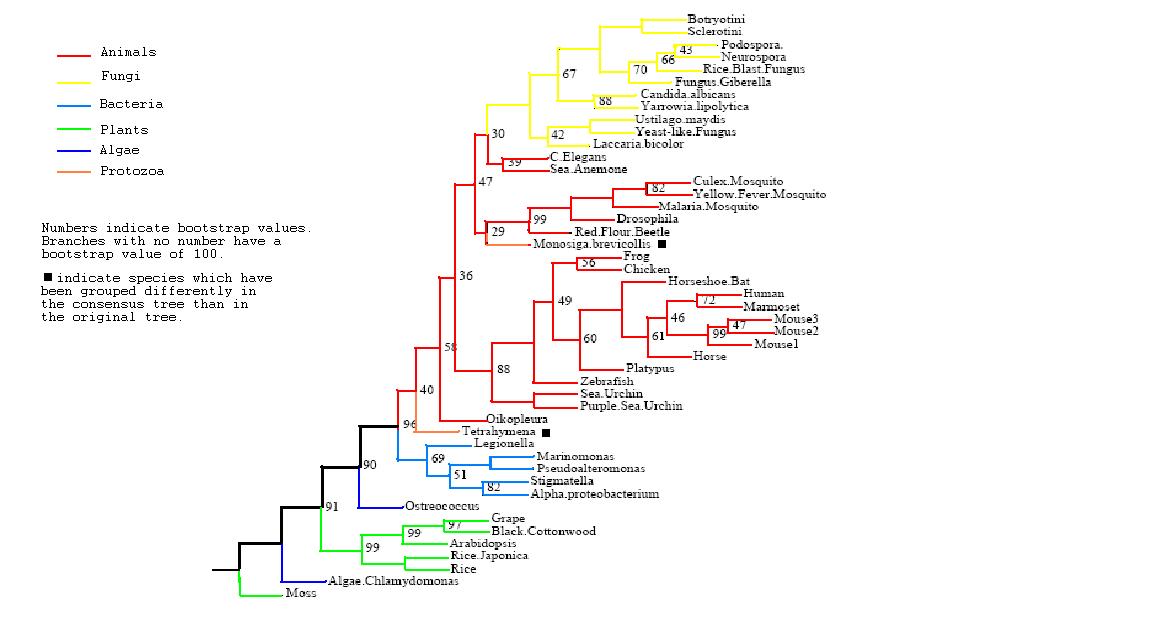
Figure 3
Rooted Consensus Tree with bootstrap values obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/
Rooted Consensus Tree with bootstrap values obtained from Phylip
Drawn using Phylodendron
http://iubio.bio.indiana.edu/treeapp/