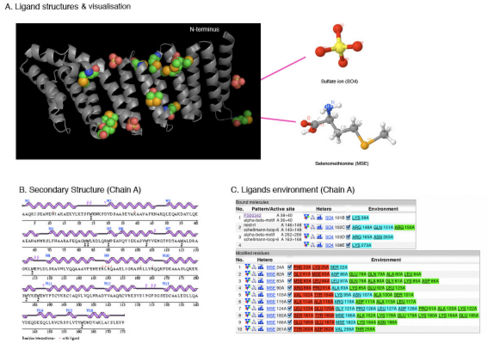
Result of SNAPG
SNAPG Structure
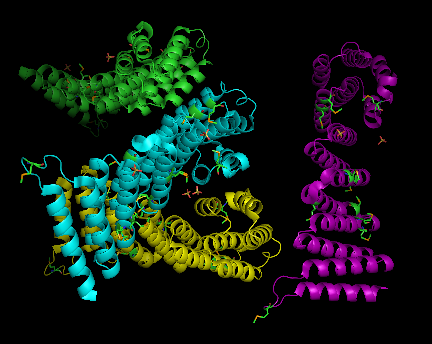
Structure architecture
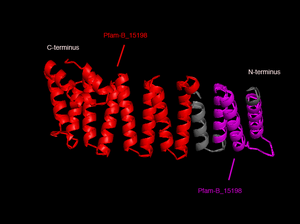
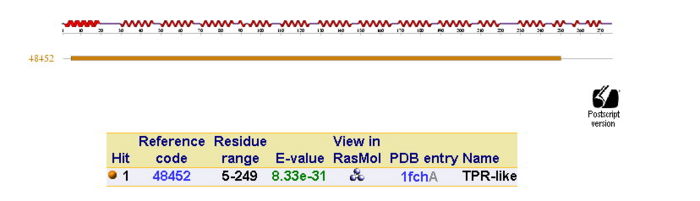
In order to analyze protein structure of SNAPG, structural comparison to known protein structure is required. An insight to SNAPG structural arrangement provides various informative data on possible protein functions and interactions with another protein and/or DNA. Based on protein families database, Pfam at Sanger, it was found that SNAPG protein matched to Pfam-B protein families and consist of 2 domains, Pfam-B_7270 (PB007270) and Pfam-B_15198 (PB015189) respectively as shown in Figure 3. Both of 2 domains appears to be associated with NSF attachment protein activity [1]. The sequence was also used against InterproScan generated by Profunc that gave an TFR (Tetratricopeptide-like helical) domain classification while that of protein family agrees with Pfam classification. (interproscan)
Structural comparison
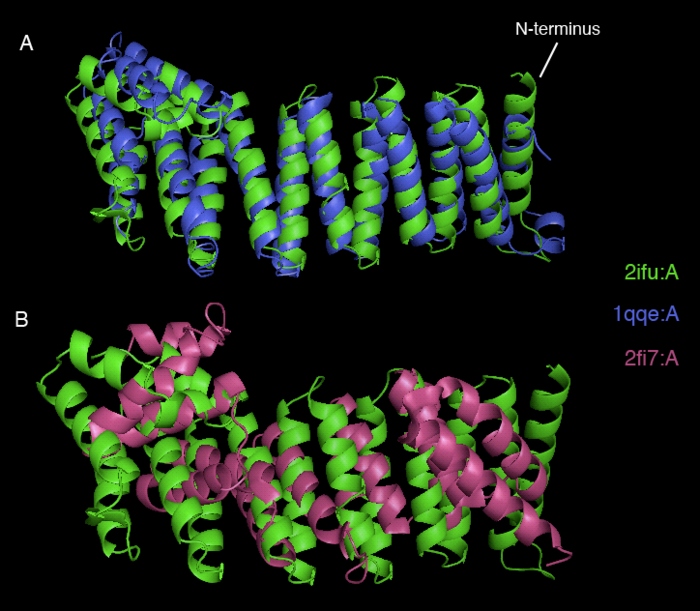
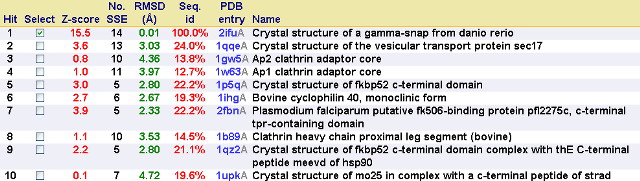
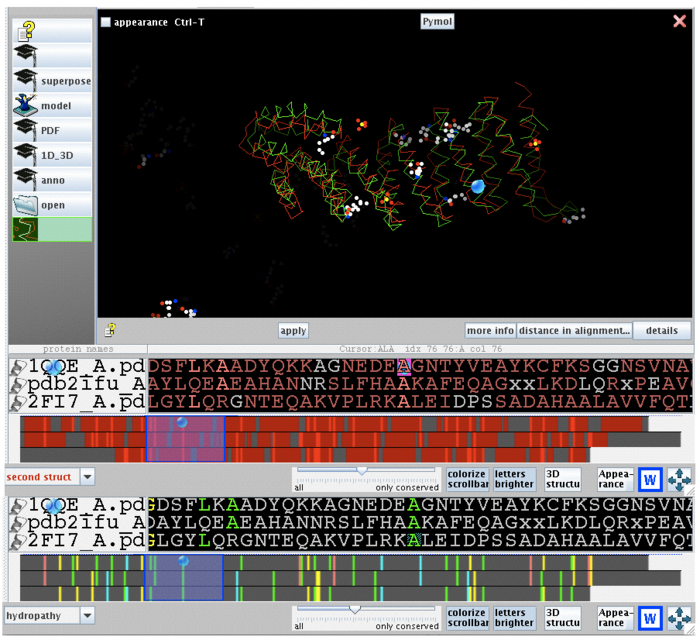
Dali webserver is one of the powerful tool to screen any protein that are structurally homologous with our query. Two structurally related proteins with highest Z-value generated by Dali server were chosen for SNAPG structure comparison analysis. These proteins were vesicular transport ptotein sec17 (1qqe) and type 4 fimbrial biogenesis protein (2f17) (refer to Table 1).
| PDB-chain | Structure | RMSD | Z-value | % identity | Protein |
|---|---|---|---|---|---|
| 2IFU-A | 0.0 | 37.8 | 100 | Endocytosis/exocytosis. Gamma-SNAP (Danio rerio) | |
| 1QQE-A | 3.4 | 23.3 | 23 | Protein binding. Vesicular transport protein sec17(yeast) | |
| 2FI7-A | 11.6 | 12.9 | 14 | Protein transport. Type 4 fimbrial biogenesis protein pili (Pseudomonas aeruginosa) |
2ifuA, 1qqeA and 2fi7 alignment
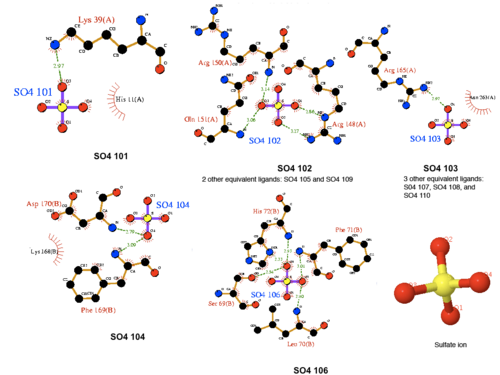
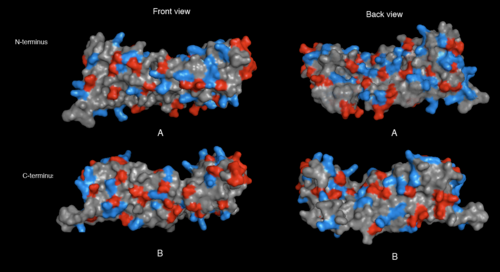
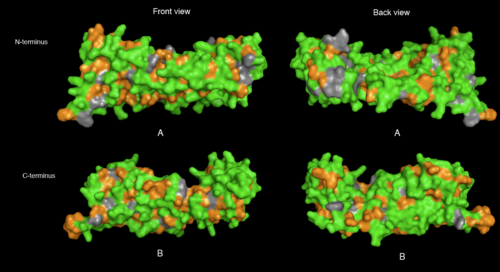
physical properties
SNAPG Function
Gene Ontology Annotation by Similarity
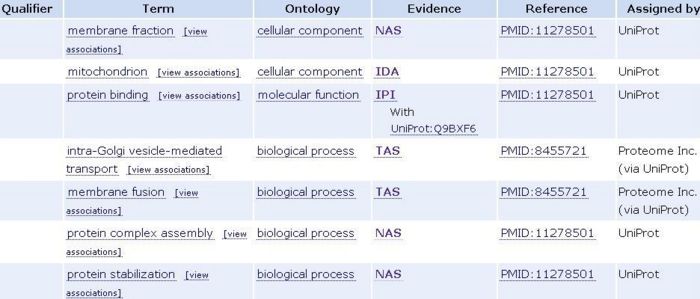
AmiGo has been used to find three structured controlled vocabularies that describe gene products of SNAPG in terms of their associated biological processes, cellular components and molecular functions. This protein function is not yet established therefore it is only by similarity (see the figure below).
Function in the context of Structure
Fold Matching Analysis by ProFunc
SSM (Krissinel and Henrick, 2004) represents several protein that have similar structure to SNAPG associated with function prediction. The most closely related structure is 1qqeA shows 24% fold identity to 2ifuA. This results could be different from DALI results (refers to Table1).
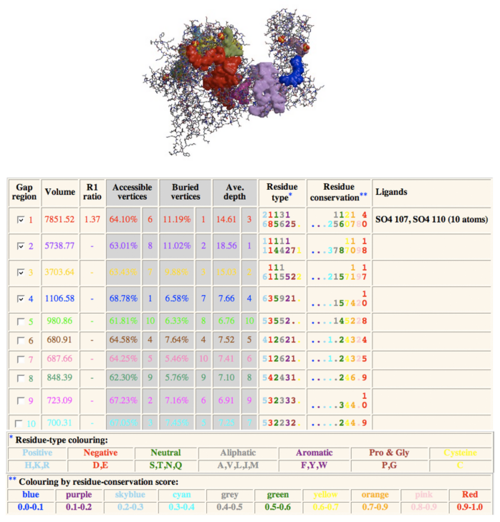
Conserved Residue Domain
Sequence Motif
Aromatic-di-Alanine (AdAR) repeat was found in NSF attachment proteins (SNAPG). Its structure is similar to that found in TPR-1 (belongs to Tetratrico peptide repeat family). Get the link
Genomic Context
Cellular Context
SNAPG Evolution
Background
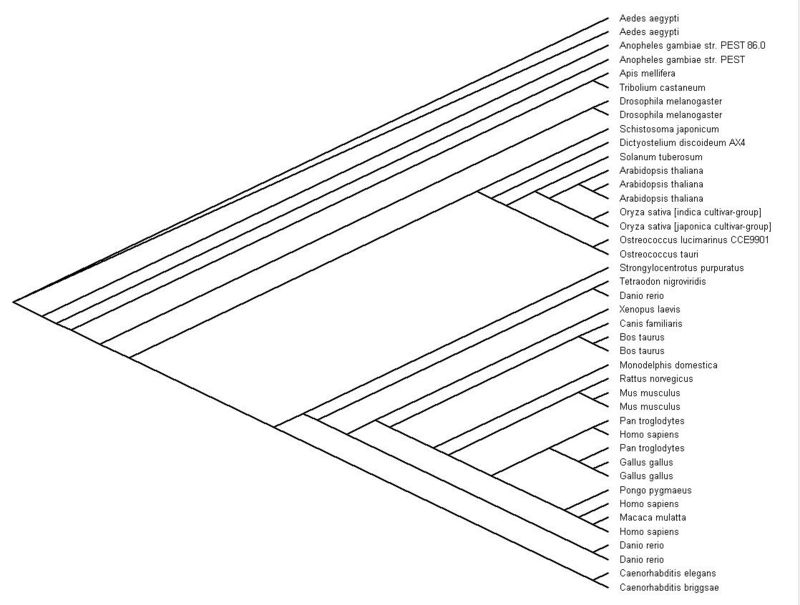
This is the cladogram treeview of SNAP-gamma protein. However, this view is not suitable enough to represent the evolutionary gap between species but this is clearly shows the species name.
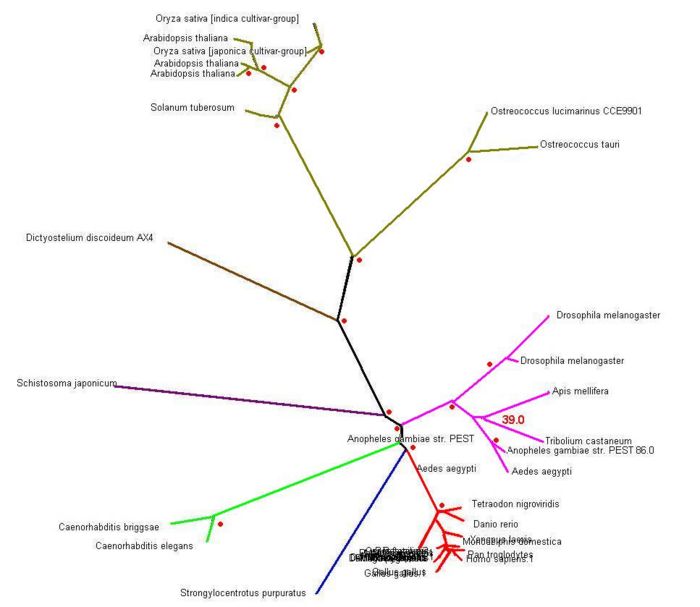
Final Tree Analysis
Legends:
- Red-dotted sign means boostrap value more than 75.00 or 75%
- any number (in red) located near branching tree indicated boostrap value of the branching tree
- Green Line: The species belongs to Plants group
- Brown Line: Amoeba group
- Purple Line: Parasite type
- Pink Line : The species belongs to Insects Group
- Light Green Line: Worms Group
- Dark Blue Line: Invertebrate
- Red Line: Vertebrate
Other version: Evolutionary analysis of the tree with radial view (for non-professional or common viewer)
Other Attributes
List IDs use as an batch entrez input
List of various alignments of protein
Back to Scientific Report of N-ethylmaleide-sensitive factor attachment protein, gamma