2i2O Protein Evolution Presentation: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
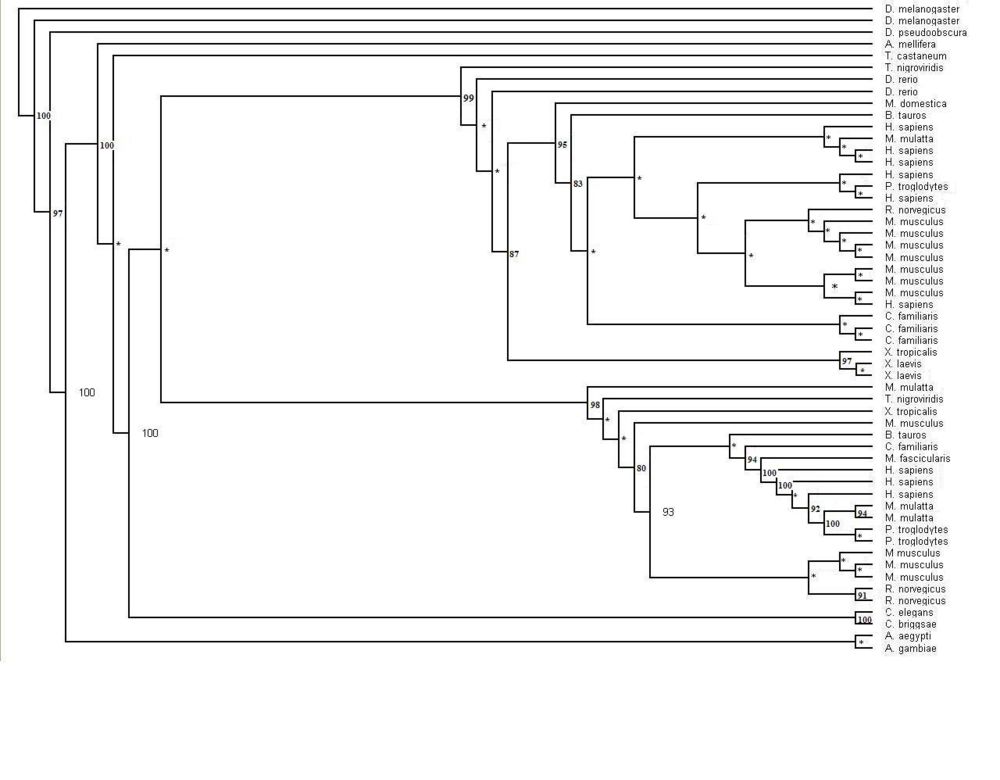
* Three programs used to construct phylogenetic tree | |||
* Blast used to find significant sequences | |||
* 54 sequences chosen because E-value under 0.01 | |||
* ClustalX used | |||
* Multiple alignment of the 55 sequences undertaken | |||
* First draft phlogenetic tree created | |||
[[Image:Protein sequence.JPG|Description]] | [[Image:Protein sequence.JPG|Description]] | ||
Revision as of 00:03, 12 June 2007
- Three programs used to construct phylogenetic tree
- Blast used to find significant sequences
- 54 sequences chosen because E-value under 0.01
- ClustalX used
- Multiple alignment of the 55 sequences undertaken
- First draft phlogenetic tree created
Figure 1.0 Protein sequence of MIF4G domain containing protein found in Homo sapiens.
Figure 1.1 Phylogenetic tree of the 55 significant sequences closest to MIFG4 protein. Bootstrapping values shown on the tree. * denotes a bootstrap value less than 75