2i2O Protein Function Presentation
Function Presentation:
• The MIF4G domain containing protein is the middle protein in the eukaryotic initiation factor 4G. eIF4G and in particular the middle domain important in the creation of a molecular bridge between eIF4G, eIF4A and RNA ribosomal units.
• Actual function is still relatively unknown
• This study was done using computational tools and databases such as ProFunc, ProKnow, LOCATE, PDB, Pfam and many others.
• Unfortunately most of the searches performed returned few or no results.
• Therefore we decided to do a comparative study. Closest related protein known to MIF4G, the MIF4G-like protein discovered in the zebra fish was used.
• Much more information about this protein was available including structural and evolutionary analysis. This assumption was made based on structural and functional comparisons between the two proteins as well as thorough literature searches.
• Database searching done using MIF4G or 2i2O, FASTA sequence or accession number.
• Where no results were returned or were not significant a second search using MIF4G-like protein or 1hu3 (or FASTA sequences etc) was done.
• Significant results from both proteins were compared for similarity.
• LOCATE 0.93 probability of the protein located in the cytoplasm
o soluble and non-secreted
o a polyadenylate binding protein-interacting protein.
o Top results all contained an ARM repeat.
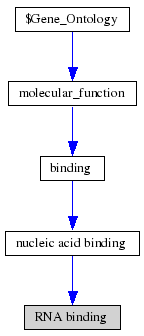
ProKnow: Identified that the likely function of MIF4G 2i2OA was RNA binding See Figure 2.4.
• Parallel to the Superfamily HMM LOCATE showed the protein to contain ARM repeats. ARM and HEAT repeats are approximately 50 residues long tandemly repeated throughout many eukaryotic proteins.
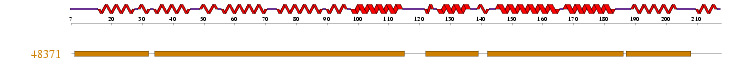
• Superfamily HMM library at residues 8-31, 34-114, 122-138, 142-185, 187-207 in the ARM repeat superfamily.
1 motif match was found to the Superfamily HMM library at residues 8-31, 34-114, 122-138, 142-185, 187-207 in the ARM repeat superfamily.
Figure 2.3: Superfamily analysis revealed 1 sequence motif in the sequence.
• Originally it was believed that ARM and HEAT repeats were similar. Recently it was found the two repeats were divergent and contain significant structural and functional differences.
o ARM repeat consists of two helices
o HEAT domain consists of three alpha helices.
• Consistent with our findings, that the domain is rich in alpha helices it provides further evidence to the function of MIF4G being the mediation of protein-protein interactions during the initiation of translation.
• Pfam identified the domain to be occurring in the eukaryotic translation initiation factor IV (eIF4) as well as in NMD2p and CBP80 (nonsense mediated mRNA decay protein 2 and nuclear cap-binding protein respectively).
• Literature showed the proteins to be structurally similar to MIF4G and also as to domains from within eIF4 domain known as HEAT domains.
o HEAT domain are a superhelical forming scaffolding matrice that is comprised of single HEAT repeat units made up of a pair of anti-parallel helices linked by a flexible loop.
o Three consecutive HEAT domains are present within eIF4 of which MIF4G is congruent with HEAT-1. The middle domain of CBP80 has been proposed to be similar to MIF4G.
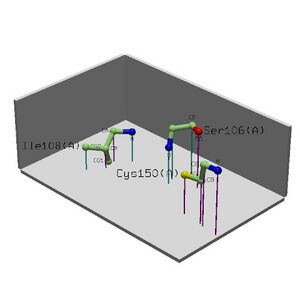
• The NEST analysis produced 3 functionally significant and conserved hits. We can infer that these structural motifs are important sites for the function of the protein.
• Combining information we can hypothesise that the structural motifs identified by NEST are important for RNA binding (the function of the protein).
Table 2.1 NEST Results
Ramachandran Solvent
Nest Score Residue range Residue region accessibility Cleft Depth cleft Res.conserv
1. 4.96 Tyr9(A)-Ile11(A) Tyr9(A) RIGHT 3.38% - - 1.00
Lys10(A) LEFT 0.52% - - 1.00
Ile11(A) - 0.31% - - 0.88
2. 3.46 Gly204(A)-Trp206(A) Gly204(A) RIGHT 0.00% - - 0.60
Gly205(A) LEFT 0.98% 2 6.70 1.00
Trp206(A) LEFT 0.00% - - 0.77
3. 2.28 Thr70(A)-Gly72(A) Thr70(A) RIGHT 0.00% - - 0.62
Asn71(A) LEFT 1.21% - - 0.68
Gly72(A) - 0.00% 4 4.42 0.54
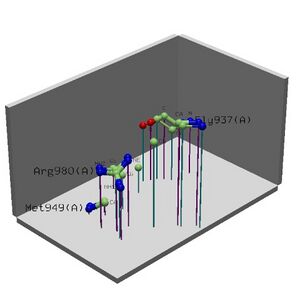
Figure 2.6 Alignment of 2i2O generated by ProFunc
• Cleft Analysis showed potential binding sites on the protein and possible residues.
• Looking collaboratively with LOCATE and ProKnow further enforce the function to be RNA binding during the initiation of translation phase.
• Literature that suggests MIF4G binds to eIF4A, an RNA helicase and RNA and is speculated to play a role in the binding of eIF4A to RNA.
Table 2.2 Cleft Analysis Results
Region R1 Accessible Buried Average Residue Residue Gap Volume 1 Ratio Vertices Vertices Depth Type Conservation Ligands 1 822.66 0.67 66.51% 2 10.40% 3 11.19 1 23662.. 1....223..2 2 1232.30 - 65.69% 3 10.80% 1 10.31 3 564644. ...1154558 3 1160.58 - 66.75% 1 8.98% 8 10.95 2 464552. ...1143548 4 931.50 - 59.62% 6 9.06% 7 8.73 5 225421. .....12257 5 827.72 - 57.62% 8 9.51% 6 8.58 6 34631.. .....22337 6 885.94 - 61.42% 4 10.04% 4 8.27 7 215421. .....12266 7 910.83 - 60.12% 5 7.92% 9 7.52 10 34263.1 ...1.11665 8 772.45 - 58.35% 7 9.64% 5 8.88 4 533.2.. ....114313 9 682.17 - 55.10% 10 5.75% 10 7.63 9 323112. .....14351 NI 502(1 atom) 10 585.14 - 57.04% 9 10.45% 2 7.93 8 2132.1. .....1317. NI 501(1 atom)
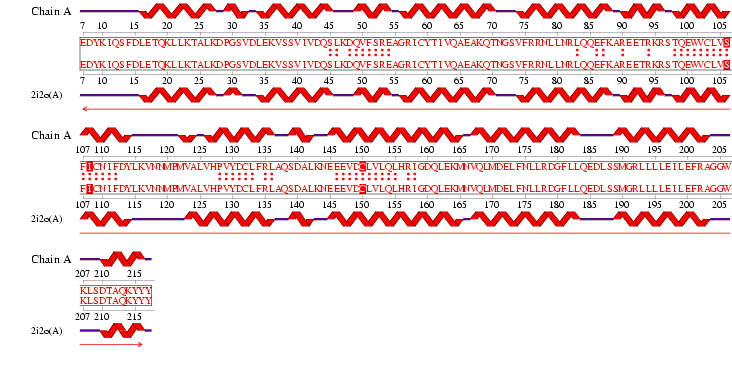
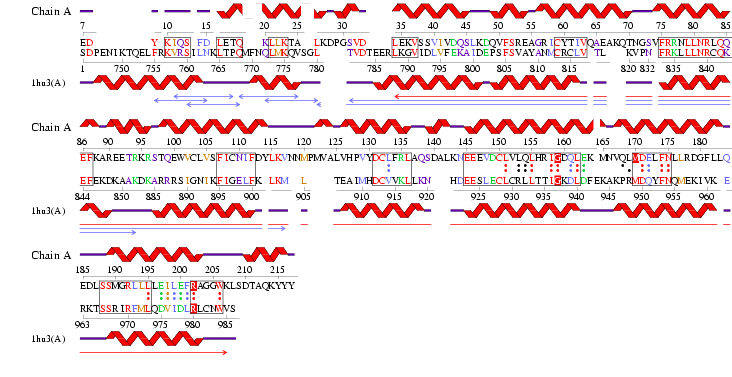
See Figure 2.7 Generated by structure comparison in PDB vs. RNA template of 1hu3 to 2i2O
• The results for 3D functional template demonstrate that 1hu3 and 2i2O are significantly structurally and functionally similar.
• Therefore it is possible to assume that the identified regions could be conserved functional regions common to both proteins.
Figure 2.9 RNA Template alignment 1hu3 vs 2i2O.
• In Conclusion based on computational analysis it can be predicted that:
o Likely function of MIF4G domain containing protein is to bind RNA
o To facilitate the simultaneous binding of another eIF4 domain eIF4A and small ribosomal units.
o This could allow the initiation of translation within eukaryotic cells.
o HEAT-domains and ARM repeats are likely to play a pivotal role in the operation of MIF4G.
• These hypotheses have been made on the assumptions that the differences between 2i2O and 1hu3 structurally and functionally are negligible.
Return to Presentation