2i2O Results
Evoltuion of MIF4G Domain Containing Protein
Results notes
First identified in Danio rerio (zebra fish)
Eukaryotic gene - derived gene
Protein conserved throughout the Metazoans (animals)
Description
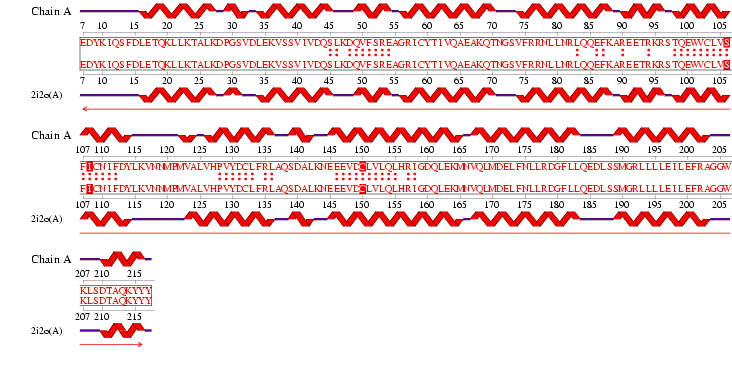
Figure 1. Protein sequence of MIF4G Domain containing protein found in Homo sapiens.
Description
Figure 2. Phylogenetic tree of the 55 significant sequences closest to eIFG4 protein. Bootstrapping values shown on the tree. * denotes a bootstrap value less than 75
Function of MIF4G Domain Containing Protein
Locate: The search using protein name MIF4G showed that the protein was most probably located in the cytoplasm of cells and is soluble and non-secreted and also identified as a polyadenylate binding protein-interacting protein. 15 results were returned, 11 of which were significant. No particular cell type was identified. When using 1hu3 as an input the results were unchanged. In addition to the location, three proteins were identified as Riken cDNA templates, being similar to the location and possible function of MIF4G, all containing an ARM repeat. These were AAH26740, AAH55812 (mouse) and AAH33579 (human, and the original sequence submitted). AAH55812 was identified as being present in a wide variety of cells including cells of the cerebellum, striatum, eye, whole brain, liver, hippocampus stem cells and kidney.
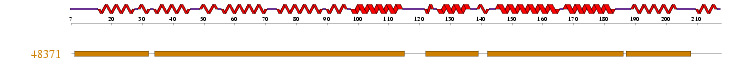
ProFunc identified results for MIF4G (2i2OA) sequence from many different databases. Interpro: found 4 motifs that were scanned and matched comparatively with PROSUTE, PRINTS, PFam-A, TIGRFAM, PROFILES and PRODOM motifs. 2 significant results from this were identified as MIF4G and eIF4G Domain. 1 motif match was found to the Superfamily HMM library at residues 8-31, 34-114, 122-138, 142-185, 187-207 in the ARM repeat superfamily.
Figure 2.0: Superfamily analysis revealed 1 sequence motif in the sequence.
Nest analysis located 3 nests in the structure containing 4.960, 3.457 and 2.284. Conservation was at 0.96, 0.79 and 0.617 respectively.
Figure 2.1: Alignment obtained from ProFunc NEST
Reverse template, structure comparisons generated 20 significant hits. (INSERT TABLE OF RESULTS) No matching structures in the PDB were found. Gene Neighbours found no matching genome locations for homologues on A or B chains. No helix-turn-helix structures were found. There were no lingand binding, DNA binding or enzyme active site templates found.
ProKnow: (picture of Gene ontology flowchart) Identified that the likely function of MIF4G 2i2OA was RNA binding as inferred by genetic interaction. This result was found using the frequency of ontology from 3D folds and the score of ontologies from 3D motifs based on conservation.
Pfam: (Picture of protein) was searched using the name MIF4G domain. The domain was identified to be occurring in NMD2p and CBP80 (nonsense mediated mRNA decay protein 2 and nuclear cap-binding protein respectively). It was found that the domain binds eIF4A, eIF3, RNA and DNA.
Structure of MIF4G Domain Containing Protein
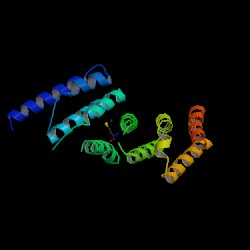
From the PDB search, the structure was revealed to be similar to “Eukaryotic Translation Initiation Factor 4G (eIF4G)” protein. The protein was isolated form Danio rerio and expressed in Escherichia coli. It is formed from two chains, with two chemical components, nickel (Ni2+) and selenomethionine (C5H11NO2Se) as additions to the protein. The NCBI Entrez search revealed the protein to be a domain of the eIF4G-like protein. Structure of MIF4G (PDB: 1hu3) Enlarge Structure of MIF4G (PDB: 1hu3)
Both Pfam and InterPro identified the protein as the Middle domain of eIF4G, termed MIF4G. MIF4G consists essentially of alpha helices and has “multiple alpha-helical repeats”. Within eIF4G, it binds to the RNA helicase eIF4A, eIF3, RNA and DNA.
The DALI server was used to identify proteins with similar structure to MIF4G-like protein from Danio rerio (PDB:2i2o). From the hits generated, three proteins (PDB:1hu3, 1uw4, 1h6k) with Z-scores 15.2, 10.9 and 10.6 respectively were selected. 1hu3 is the middle domain of human “Eukaryotic Translation Initiation Factor 4G (eIF4G)”. 1uw4 is an mRNA decay factor and 1h6k is the human nuclear cap binding protein complex (CBC).
A CE structural comparison of 1hu3 with the zebrafish putative MIF4G revealed much similarity in folding and protein component. 4 alpha-helices folded in the same orientation can be identified on each protein. The generated figure shows a superimposed image of both proteins, hence suggesting an overall similarity in structure. The N-terminal of 1h6k is highly similar in fold and orientation as MIF4G. This implies a possibility that MIF4G protein is a region, or even an active domain, near the N-terminal of the CBC.
Return to Scientific Report