2i2O Results
Evoltuion of MIF4G Domain Containing Protein
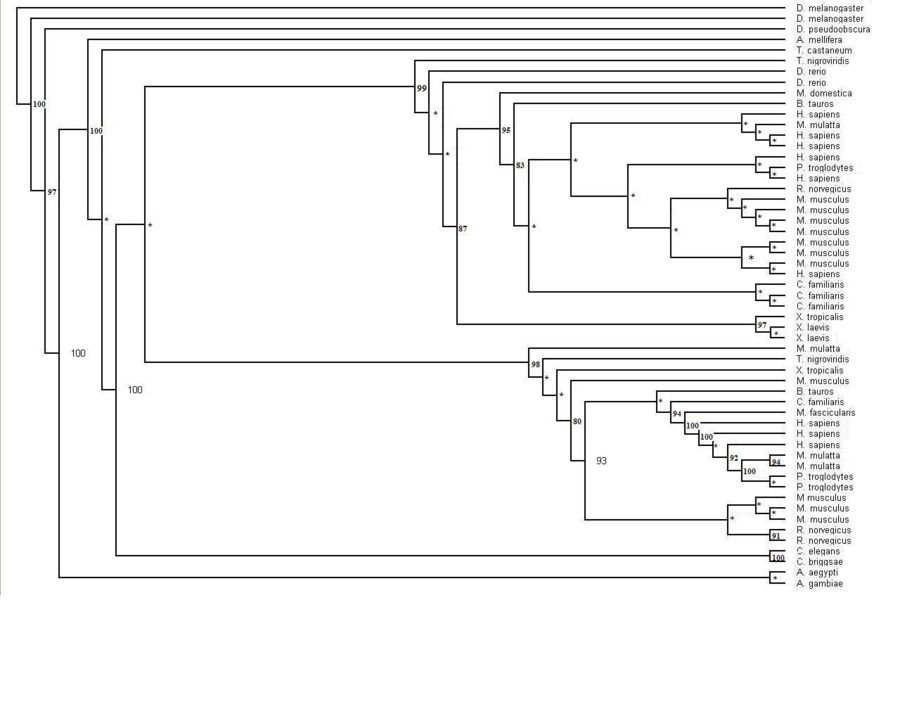
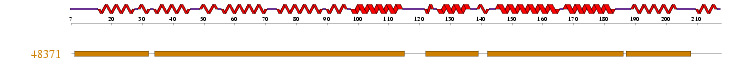
The human sequence of MIF4G (Figure 1.0) was also blasted in the NCBI website. The website identified that this protein was first isolated from zebrafish Danio rerio. Several related sequences of MIF4G were examined for specific information. The information found suggested that this particular protein was highly conserved throughout the Metazoans (animals). The multiple alignment sequence that was conducted indicated several areas of high conservation within the MIF4G protein (Fig 1.1). Although early research suggests that this particular domain containing protein is part of the eukaryotic initiation translation factor, no sequences from the two other main eukaryotic families, plants and fungi (Figure 1.2). The phylogenetic tree with the bootstrap values incorporated has several organisms that have more than one sequences displayed on the tree. The main phylogenetic tree for the MIF4G domain containing protein is split in to two smaller trees within the major tree. The middle sequences within these two trees consist of sequences from mammals.
Figure 1.0 Protein sequence of MIF4G domain containing protein found in Homo sapiens.
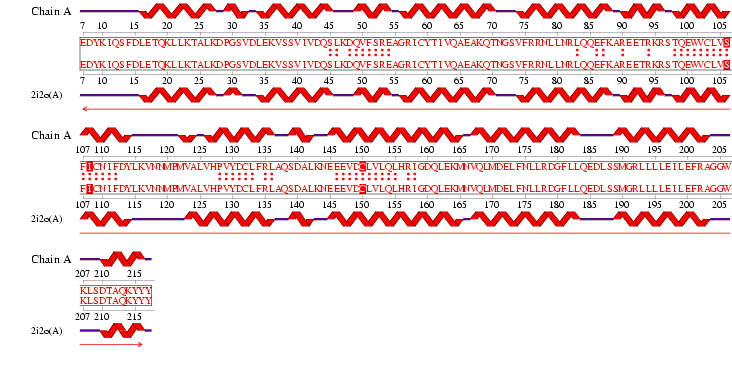
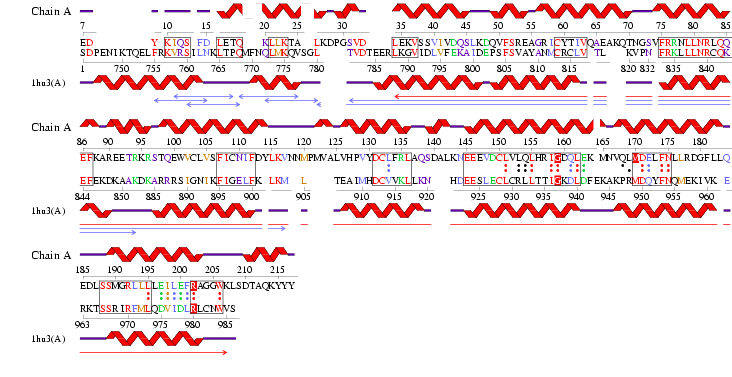
Figure 1.1 Multiple alignment sequence of the MIF4G domain containing protein. The top sequence is the human sequence of the MIF4G protein and the focus of this report.
Figure 1.2 Phylogenetic tree of the 55 significant sequences closest to MIFG4 protein. Bootstrapping values shown on the tree. * denotes a bootstrap value less than 75
Function of MIF4G Domain Containing Protein
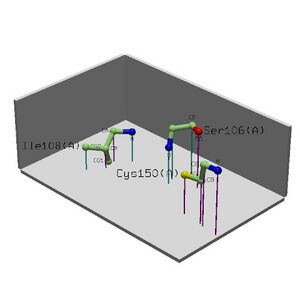
Locate: The search using protein name MIF4G showed that the protein was most probably located in the cytoplasm of cells and is soluble and non-secreted and also identified as a polyadenylate binding protein-interacting protein. 15 results were returned, 11 of which were significant. No particular cell type was identified. When using 1hu3 as an input the results were unchanged. In addition to the location, three proteins were identified as Riken cDNA templates, being similar to the location and possible function of MIF4G, all containing an ARM repeat. These were AAH26740, AAH55812 (mouse) and AAH33579 (human, and the original sequence submitted). AAH55812 was identified as being present in a wide variety of cells including cells of the cerebellum, striatum, eye, whole brain, liver, hippocampus stem cells and kidney.
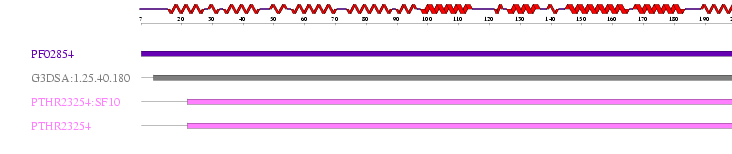
ProFunc identified results for MIF4G (2i2OA) sequence from many different databases. Interpro: found 4 motifs that were scanned and matched comparatively with PROSUTE, PRINTS, PFam-A, TIGRFAM, PROFILES and PRODOM motifs. 2 significant results from this were identified as MIF4G and eIF4G Domain.
Table 2.0 InterPro Results
Hit Scan Reference code Residue range Motif name 1. HMMPfam PF02854 7-205 MIF4G 2. G3DSA:1.25.40.180 11-206 no description 3. HMMPanther PTHR23254:SF10 22-217 AD023 PROTEIN 4. HMMPanther PTHR23254 22-217 EIF4G DOMAIN PROTEIN
Figure 2.2 Results generated by Interpro
1 motif match was found to the Superfamily HMM library at residues 8-31, 34-114, 122-138, 142-185, 187-207 in the ARM repeat superfamily.
Figure 2.3: Superfamily analysis revealed 1 sequence motif in the sequence.
Nest analysis located 3 nests in the structure containing 4.960, 3.457 and 2.284. Conservation was at 0.96, 0.79 and 0.617 respectively.
Figure 2.6: Alignment obtained from ProFunc NEST
Results generated by NEST are as follows
Table 2.1 NEST Results
Ramachandran Solvent
Nest Score Residue range Residue region accessibility Cleft Depth in cleft Residue conservation
1. 4.96 Tyr9(A)-Ile11(A) Tyr9(A) RIGHT 3.38% - - 1.00
Lys10(A) LEFT 0.52% - - 1.00
Ile11(A) - 0.31% - - 0.88
2. 3.46 Gly204(A)-Trp206(A) Gly204(A) RIGHT 0.00% - - 0.60
Gly205(A) LEFT 0.98% 2 6.70 1.00
Trp206(A) LEFT 0.00% - - 0.77
3. 2.28 Thr70(A)-Gly72(A) Thr70(A) RIGHT 0.00% - - 0.62
Asn71(A) LEFT 1.21% - - 0.68
Gly72(A) - 0.00% 4 4.42 0.54
Cleft Analysis generated within ProFunc found 10 gap regions as pictured in Figure 2.2.
Table 2.2 Cleft Analysis Results
Region R1 Accessible Buried Average Residue Residue Gap Volume 1 Ratio Vertices Vertices Depth Type Conservation Ligands 1 822.66 0.67 66.51% 2 10.40% 3 11.19 1 23662.. 1....223..2 2 1232.30 - 65.69% 3 10.80% 1 10.31 3 564644. ...1154558 3 1160.58 - 66.75% 1 8.98% 8 10.95 2 464552. ...1143548 4 931.50 - 59.62% 6 9.06% 7 8.73 5 225421. .....12257 5 827.72 - 57.62% 8 9.51% 6 8.58 6 34631.. .....22337 6 885.94 - 61.42% 4 10.04% 4 8.27 7 215421. .....12266 7 910.83 - 60.12% 5 7.92% 9 7.52 10 34263.1 ...1.11665 8 772.45 - 58.35% 7 9.64% 5 8.88 4 533.2.. ....114313 9 682.17 - 55.10% 10 5.75% 10 7.63 9 323112. .....14351 NI 502(1 atom) 10 585.14 - 57.04% 9 10.45% 2 7.93 8 2132.1. .....1317. NI 501(1 atom)
PDB Database found 4 significant matching sequences. These were found by submitting FASTA sequence of 2i2O
Table 2.3 PDB Results
PDB code %-tage id Overlap Name 1. 2i2o(A) 100.000 100 Crystal structure of an eif4g-like protein from danio rerio 2. 1hu3(A) 25.287 59 Middle domain of human eif4gii 3. 1vkh(A) 20.792 57 Crystal structure of putative serine hydrolase (ydr428c) from saccharomyces cerevisiae at 1.85 a resolution 4. 1suu(A) 28.125 56 Structure of DNA gyrase a c-terminal domain
Reverse template, structure comparisons generated 20 significant hits. Reverse Template search results:
2 significant hits were produced by the ProFunc database for reverse template search.
- 1 E-value of 0.00E+00 showed 960.00 similarity with 100 sequence identity and overlap. Structural similarity was 99.5%. This was identified as our protein 2i2O.
- 2 E-value of 1.98E-04 showed 342.91 similarity with 25% seuqnece identity but 98.4% structural similarity. This was found to be 1hu3 - MIF4G like protein from Danio Renio.
No matching structures in the PDB were found. Gene Neighbours found no matching genome locations for homologues on A or B chains. No helix-turn-helix structures were found. There were no lingand binding, DNA binding or enzyme active site templates found.
Figure 2.5: 3D functional template searches - Reverese template comparison vs PDB structures. 1hu3 vs 2i2O.
ProKnow: Identified that the likely function of MIF4G 2i2OA was RNA binding as inferred by genetic interaction. This result was found using the frequency of ontology from 3D folds and the score of ontologies from 3D motifs based on conservation.
Pfam: was searched using the name MIF4G domain. The domain was identified to be occurring in NMD2p and CBP80 (nonsense mediated mRNA decay protein 2 and nuclear cap-binding protein respectively). It was found that the domain binds eIF4A, eIF3, RNA and DNA.
Structure of MIF4G Domain Containing Protein
From the PDB search, the structure was revealed to be similar to “Eukaryotic Translation Initiation Factor 4G (eIF4G)” protein. The protein was isolated from Danio rerio and expressed in Escherichia coli. It is formed from two chains, with two chemical components, nickel (Ni2+) and selenomethionine (C5H11NO2Se) as additions to the protein. The NCBI Entrez search revealed the protein to be a domain of the eIF4G-like protein.
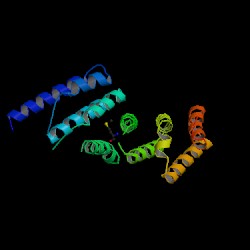
Both Pfam and InterPro identified the protein as the Middle domain of eIF4G, termed MIF4G. MIF4G consists essentially of alpha helices and has “multiple alpha-helical repeats”. Within eIF4G, it binds to the RNA helicase eIF4A, eIF3, RNA and DNA.
The DALI server was used to identify proteins with similar structure to MIF4G-like protein from Danio rerio (PDB:2i2o). From the hits generated, three proteins (PDB:1hu3, 1uw4, 1h6k) with Z-scores 15.2, 10.9 and 10.6 respectively were selected. 1hu3 is the middle domain of human “Eukaryotic Translation Initiation Factor 4G (eIF4G)”. 1uw4 is an mRNA decay factor and 1h6k is the human nuclear cap binding protein complex (CBC).
Table 3.0 DALI Results
- SUMMARY: PDB/chain identifiers and structural alignment statistics
NR. STRID1 STRID2 Z RMSD LALI LSEQ2 %IDE REVERS PERMUT NFRAG TOPO PROTEIN 1: 3028-A 2i2o-A 37.1 0.0 206 206 100 0 0 1 S STRUCTURAL GENOMICS, UNKNOWN FUNCTION eif4g-like prote 2: 3028-A 1hu3-A 15.2 2.7 164 204 23 0 0 12 S TRANSLATION eif4gii fragment (eukaryotic initiation f 3: 3028-A 1uw4-B 10.9 3.6 164 247 9 0 0 12 S NONSENSE MEDIATED MRNA DECAY PROTEIN regulator of nons 4: 3028-A 1h6k-A 10.6 3.9 175 728 11 0 0 11 S NUCLEAR PROTEIN cbp80 fragment (ncbp 80 kda subunit, 5: 3028-A 2db0-A 8.1 3.3 148 239 14 0 0 13 S PROTEIN BINDING 253aa long hypothetical protein (hypot 6: 3028-A 1b3u-A 8.0 27.7 150 588 13 0 0 13 S SCAFFOLD PROTEIN protein phosphatase pp2a fragment
A CE structural comparison of 1hu3 with the zebrafish putative MIF4G revealed much similarity in folding and protein component. 4 alpha-helices folded in the same orientation can be identified on each protein. The generated figure shows a superimposed image of both proteins, hence suggesting an overall similarity in structure. The N-terminal of 1h6k is highly similar in fold and orientation as MIF4G. This implies a possibility that MIF4G protein is a region, or even an active domain, near the N-terminal of the CBC.
Return to Scientific Report