3b5q Evolution: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
|||
| Line 277: | Line 277: | ||
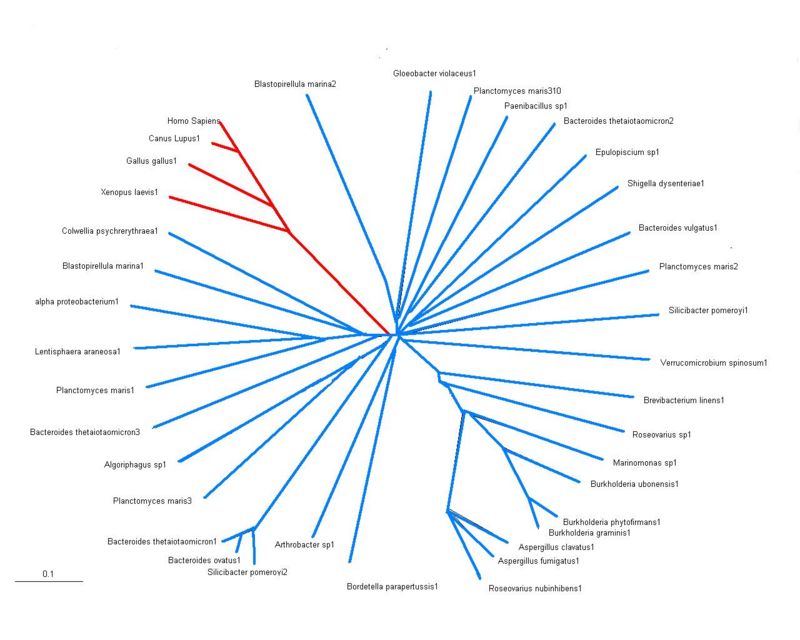
[[Image:treeA.jpg|right|thumb|800px|'''Figure 2''' | [[Image:treeA.jpg|right|thumb|800px|'''Figure 2'''<Br>Evolutionary tree of proteins showing sequence homology with arylsulfatase K, red represents eukaryotes and blue represents bacteria]]<Br> | ||
[[Image:bootstrap tree_final.jpg|right|thumb|800px|'''Figure 3''' | [[Image:bootstrap tree_final.jpg|right|thumb|800px|'''Figure 3'''<Br>Phylogeny rooted tree showing bootstrap values]]<Br> | ||
Revision as of 07:37, 9 June 2008
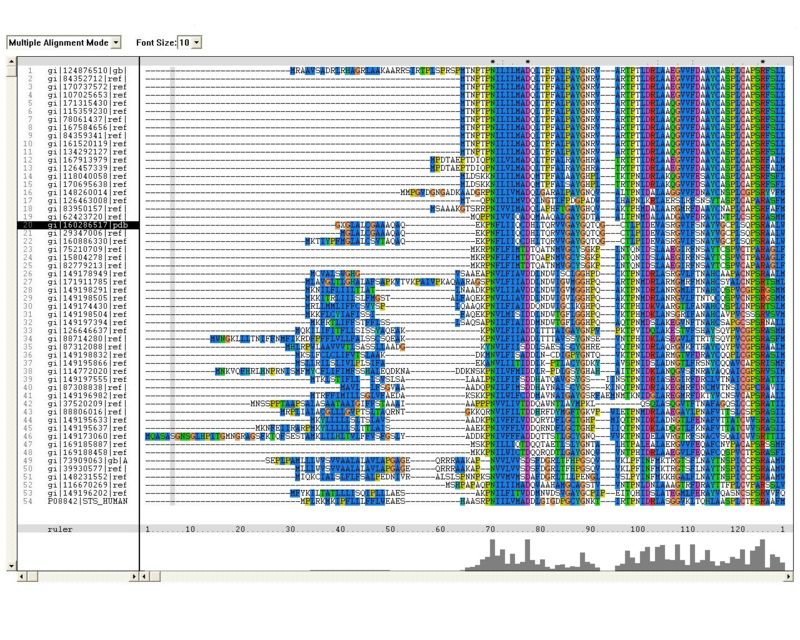
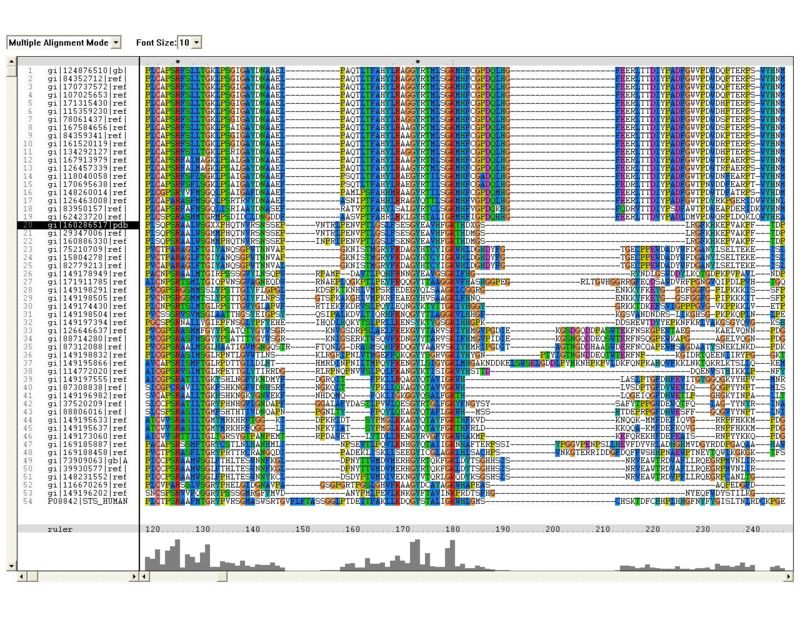
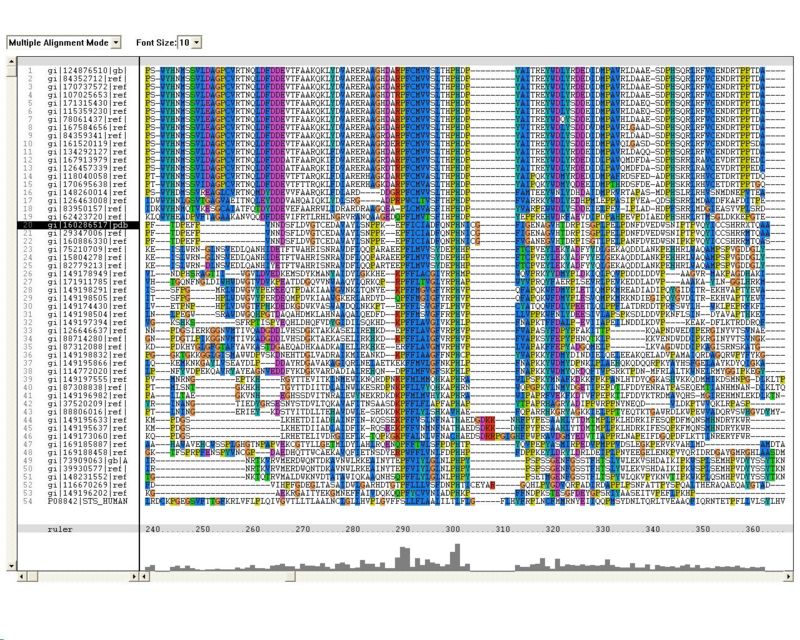
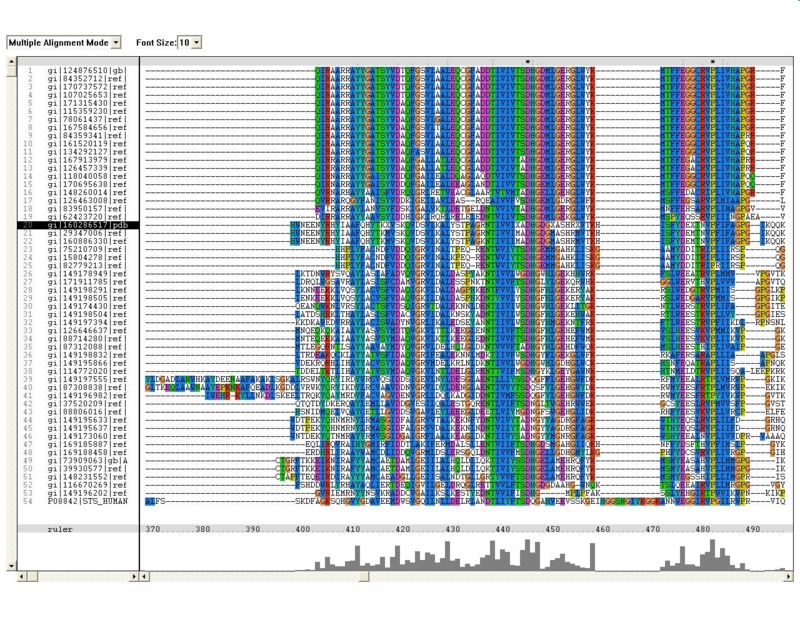
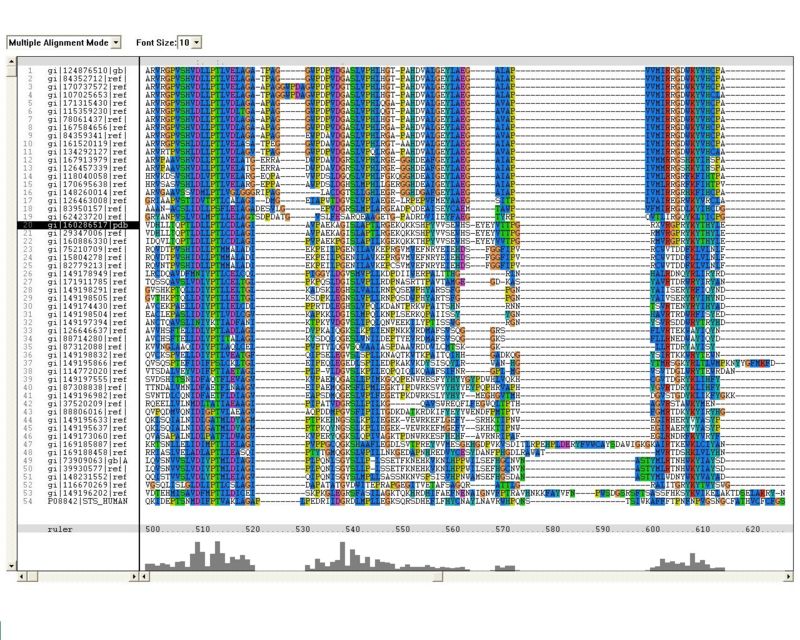
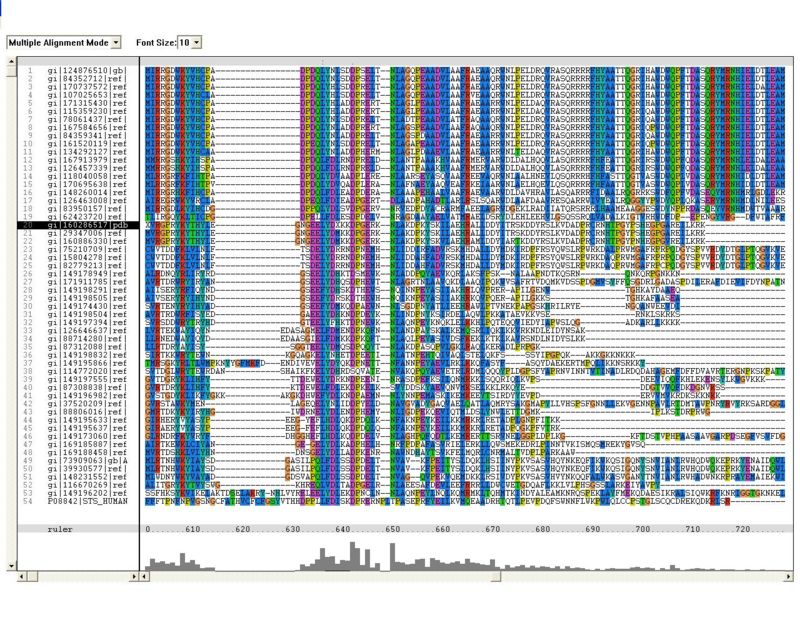
Sequence Homology
Multiple sequence alignment (MSA)
Proteins, Organisms names & Sequences
Evolutionary Tree
Sequence homology of arylsulfatases
Click here to go Back