3b5q Function: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
|||
| Line 1: | Line 1: | ||
Protein data base profile characteris arylsulfatase asa hydrolase and a sulfatase. A hydrolase is an enzyme which hydrolyses a chemical bond. The general reaction can be shown as follows. | |||
Group of ydrolaes are named as '''EC 3''' in enzyme classification. | |||
== 1. Function of sulfatases == | == 1. Function of sulfatases == | ||
Revision as of 02:56, 21 May 2008
Protein data base profile characteris arylsulfatase asa hydrolase and a sulfatase. A hydrolase is an enzyme which hydrolyses a chemical bond. The general reaction can be shown as follows.
Group of ydrolaes are named as EC 3 in enzyme classification.
1. Function of sulfatases
Sulfatases are enzymes,which hydrolyse sulfate ester bonds of substrates. Most of the family members has shown to contain a highly conserved cystine residue and a bivalent metal binding site.
2. Functional site
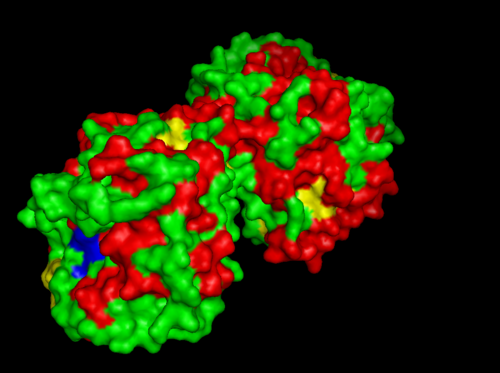
MSA data revealed some conserved residues on the sequence. They were mapped on the three dimantional structure.
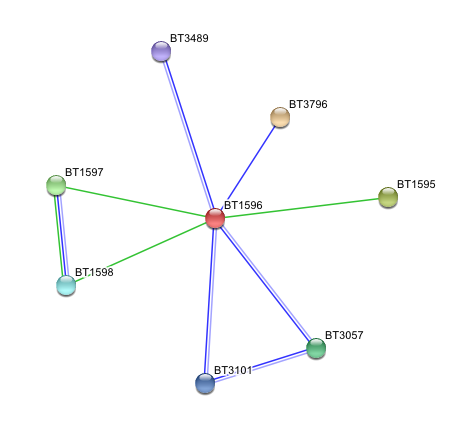
3. Arylsulfatase K (BT1596) Interactions with other proteins
Input Protein
- BT1596 Putative sulfatase yidJ (481 aa) of Bacteroides thetaiotaomicron.
Predicted Functional Partners
- BT3796: Putative secreted sulfatase ydeN (518 aa).
- BT1595 Transcription termination factor rho (722 aa).
- BT1597 Two-component system sensor histidine kinase (539 aa).
- BT3057 N-acetylgalactosamine-6-sulfatase (508 aa).
- BT1598 Putative two-component system sensor histidine (655 aa).
- BT3101 N-sulphoglucosamine sulphohydrolase (455 aa).
- BT3489 Arylsulfatase B {UniProtKB/TrEMBL-Q8A219} (458 aa).
Click here to go Back