|
|
| Line 25: |
Line 25: |
|
| |
|
|
| |
|
| '''Sequence'''
| |
|
| |
| >gi|160286517|pdb|3B5Q|A Chain A, Crystal Structure Of A Putative Sulfatase (Np_810509.1) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.40 A Resolution
| |
| GXGLALCGAAAQAQEKPNFLIIQCDHLTQRVVGAYGQTQGCTLPIDEVASRGVIFSNAYVGCPLSQPSRA
| |
| ALWSGXXPHQTNVRSNSSEPVNTRLPENVPTLGSLFSESGYEAVHFGKTHDXGSLRGFKHKEPVAKPFTD
| |
| PEFPVNNDSFLDVGTCEDAVAYLSNPPKEPFICIADFQNPHNICGFIGENAGVHTDRPISGPLPELPDNF
| |
| DVEDWSNIPTPVQYICCSHRRXTQAAHWNEENYRHYIAAFQHYTKXVSKQVDSVLKALYSTPAGRNTIVV
| |
| IXADHGDGXASHRXVTKHISFYDEXTNVPFIFAGPGIKQQKKPVDHLLTQPTLDLLPTLCDLAGIAVPAE
| |
| KAGISLAPTLRGEKQKKSHPYVVSEWHSEYEYVTTPGRXVRGPRYKYTHYLEGNGEELYDXKKDPGERKN
| |
| LAKDPKYSKILAEHRALLDDYITRSKDDYRSLKVDADPRCRNHTPGYPSHEGPGAREILKRK
| |
|
| |
| == Links ==
| |
|
| |
| [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&val=160286517 Arylsulfatase sequence]
| |
|
| |
| [http://www.ncbi.nlm.nih.gov/pubmed/17558559?ordinalpos=2&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum Human sulfatases: a structural perspective to catalysis]
| |
|
| |
| [http://www.ncbi.nlm.nih.gov/pubmed/10212197?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DiscoveryPanel.Pubmed_Discovery_RA&linkpos=4&log$=relatedarticles&logdbfrom=pubmed Amino acid residues forming the active site of arylsulfatase A. Role in catalytic activity and substrate binding]
| |
|
| |
| [http://www.rcsb.org/pdb/explore/explore.do?structureId=3b5q Protein structure in PDB]
| |
|
| |
| [http://www.ncbi.nlm.nih.gov/pubmed/9521684?dopt=Abstract Function of sulfatases; Arylsulfatase A]
| |
|
| |
|
| == Crystal Structure == | | == Crystal Structure == |
Revision as of 23:43, 25 May 2008
Materials & Methods
1) Protein structural search on PyMOL, NCBI Entrez, InterPro and PDB databases
2) Comparing protein search with DALI server to identify similiar structures
Results
PDB
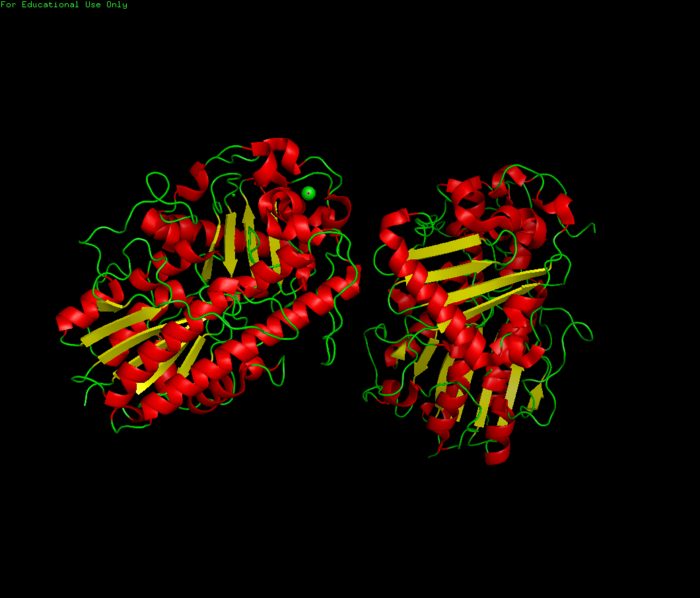
Figure 1.0 Structure of 3b5q
Discussion
Crystal Structure
Click here to go Back