ATP binding domain 4 Evolution
METHODS
The protein that is used in this research is a putative n-type ATP pyrophosphatase from Pyrococcus furiosus. This protein is predicted to be similar to ATP binding domain 4. From ‘Target Blast and Symatlas Table’, a Blastp search against the NCBI non-redundant protein database can be conducted for both sequences from Pyrococcus furiosus ATP pyrophosphatase Macaca mulatta ATP binding domain 4. Sequences with small E value were selected and Fasta format of selected sequences were retrieved. Using these sequences, a multiple sequence alignment was conducted by using ClustalX. A phylogenetic tree can be constructed based on the best multiple sequence alignment so that the evolutionary relationship between species can be observed. From the phylogenetic tree, bootstrap was performed with 100 replicates thus bootstrap values which indicate the branch length of the tree can be calculated. STRING database
RESULTS
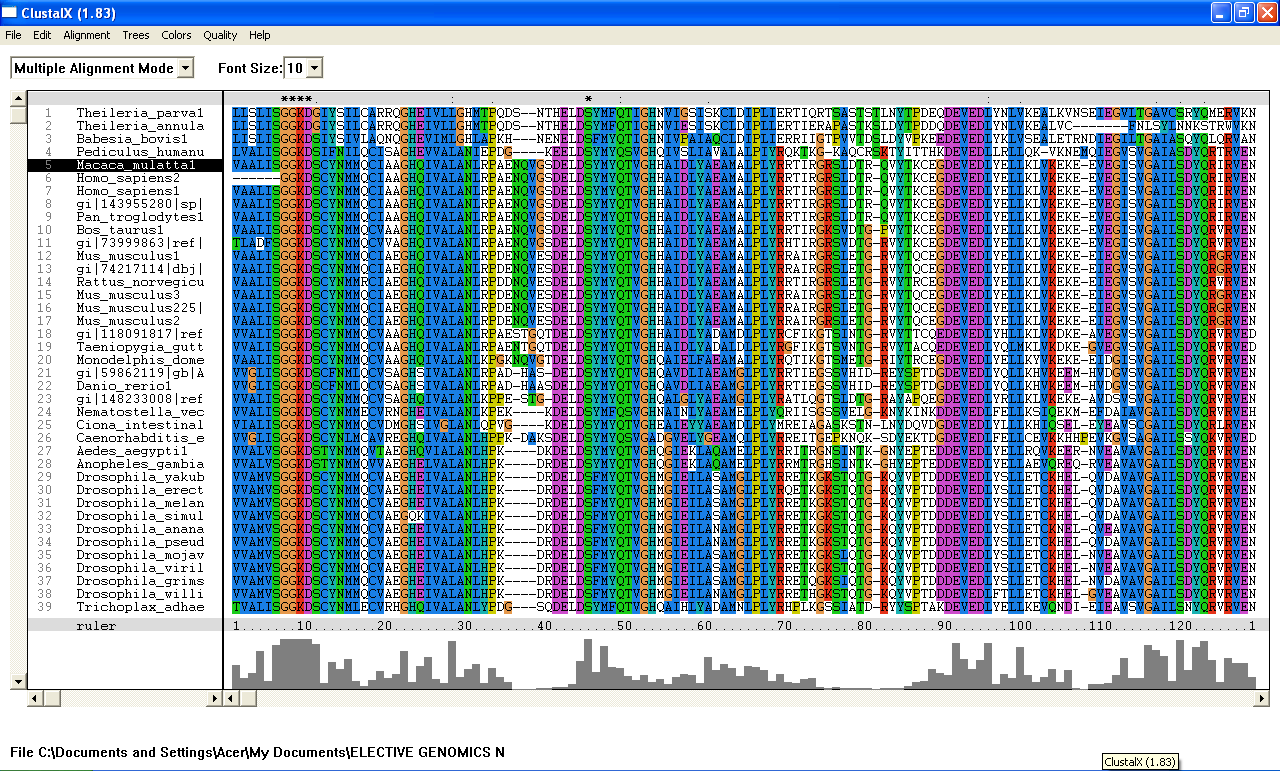
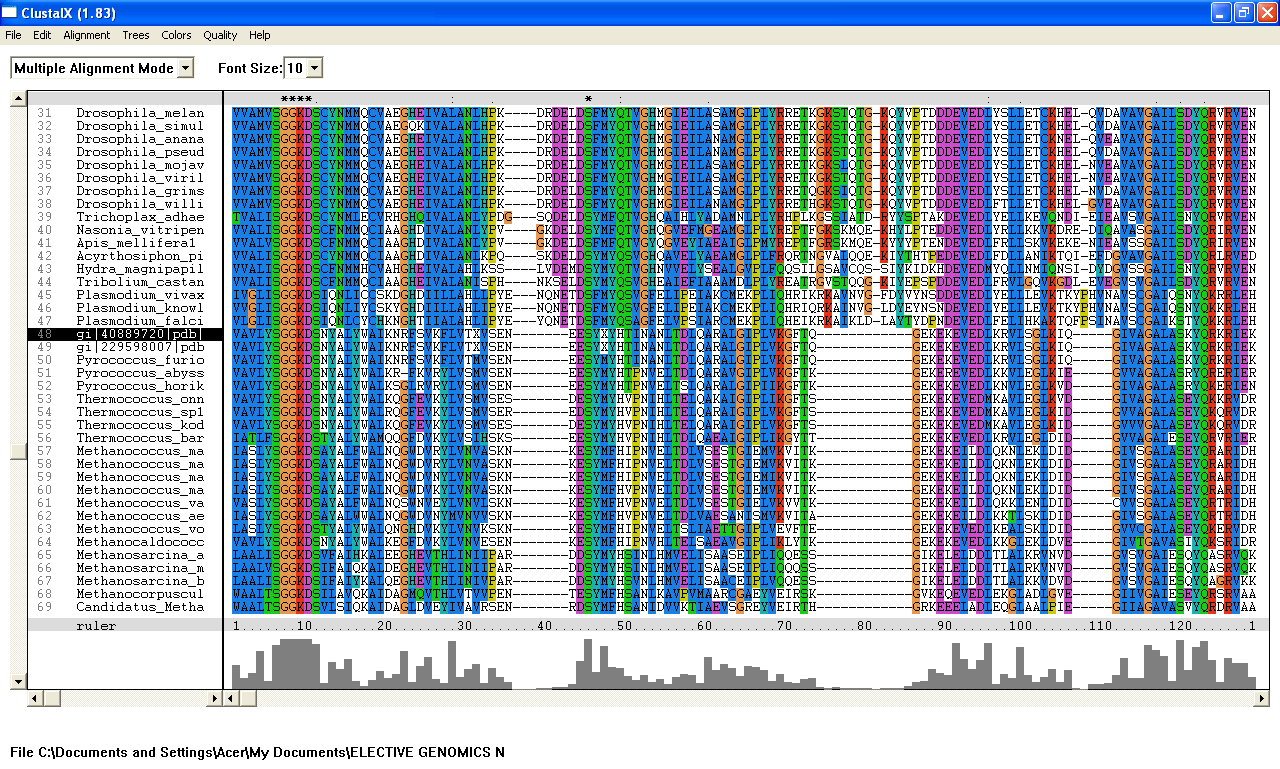
From the Blastp search, it was found that the E value for both query sequence i.e; 1ru8A and the human ortholog, are very low. The highest E-value for 1ru8A and the human ortholog are 2e-25 and 1e-49 respectively which are very low. Therefore, all 200 sequences were aligned by using ClustalX. From the multiple sequence alignment, it was found that only one conserved region presence in the sequences. Therefore, only extremely low E values were re-selected and unrelated sequences were ignored and deleted. Based on these new re-selected sequences, it was found that there are 7 conserved residues......
Multiple sequence alignment-ClustalX
DISCUSSION
Multiple sequence alignment
From the multiple sequence alignment, there are seven conserved regions found in all species from Domain Eukaryote and Archaea (based on Blastp search) but only three conserved residues which are significant to the function and structure of the protein since these residues are identified to be conserved amino acid sequence motif for P-loop of nucleotide binding domains which might be important in phosphate binding. These conserved residues are Serine-103, Glysine-104 and Glysine-105. Since this motif presence in uncharacterized ATP pyrophosphatase domain, the motif is called PP motif and can be written as S-G (2)-K-D-[GS]. This PP-loop motif is a modified version of the P-loop of nucleotide binding domain that is involved in phosphate binding. However, based on the multiple sequence alignment, the amino acids for PP-loop that are conserved across all species in eukaryote and archaea are Glysine-104 and Glysine-105. Substitution of Serine with Threonine in Pyrobaculum arsenaticum, Pyrobaculum calidifontis, Thermoproteus neutrophilis, Pyrobaculum islandicum and Staphlothermus marinus, are conserved since both amino acids are polar uncharged amino acid, suggesting that this residue is also important. However, this residue is not conserved in ATPBD4 in Homo sapiens which is only encoded by 259 amino acid sequences and the sequence started from Glysince-104. Other sequences which are conserved across all species are Lysine-106 and Aspartic acid-107 where Aspartic acid-107 is also a part of PP motif. In fact, amino acid in residue 108 is also one of the important residues in PP motif. This residue is conserved across all species except for Theileria parva and Theileria annulata where Serine is substituted with Glysine. Blastp search revealed the very low E value from many sequences from different species. These sequences appear to be encoding different group of proteins such as ATP pyrophosphatase, ATP-binding protein, ATPase and endoribonuclease. Although these proteins are responsible for different functions, the similarities of sequence between the proteins across species are very high (as indicated by E value) and the sequences which are conserved are a part of PP motif, suggesting that these proteins descend from a common ancestral sequence and therefore are paralogs. Such phenomenon could be as the result of duplication of genes within a genome.
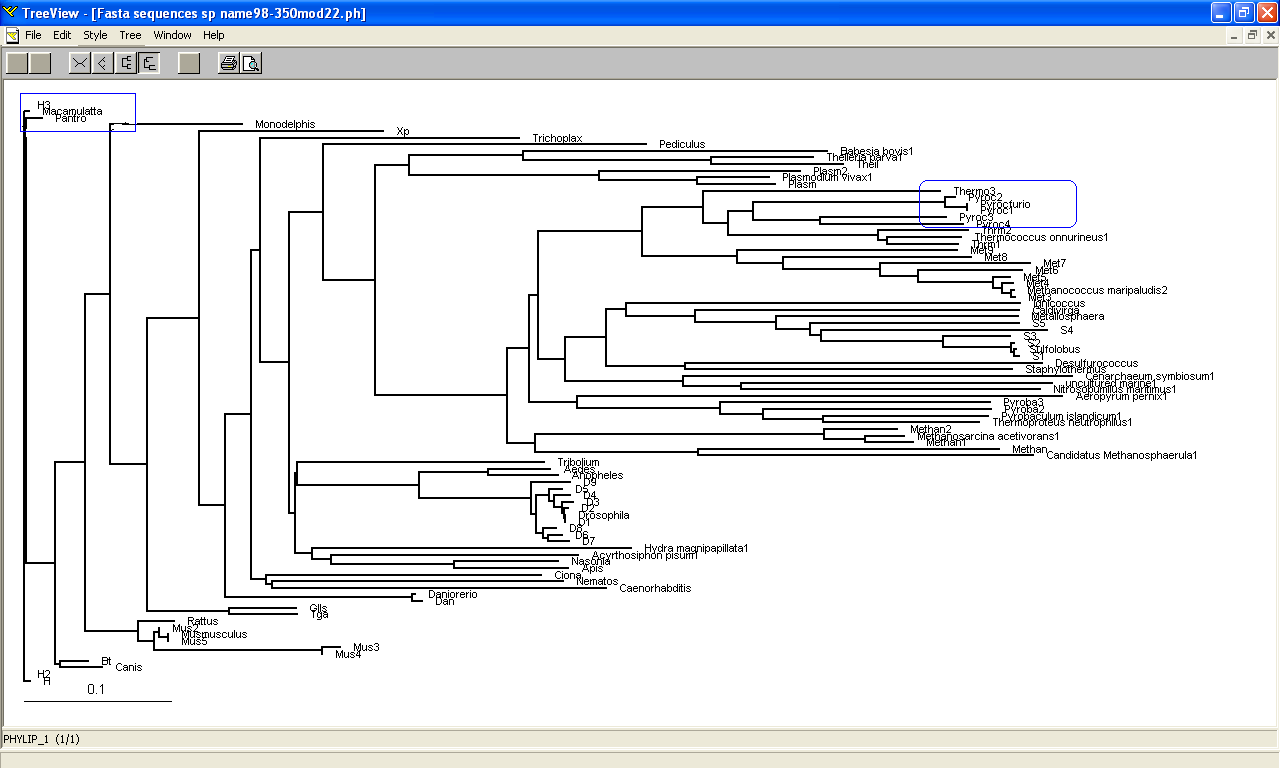
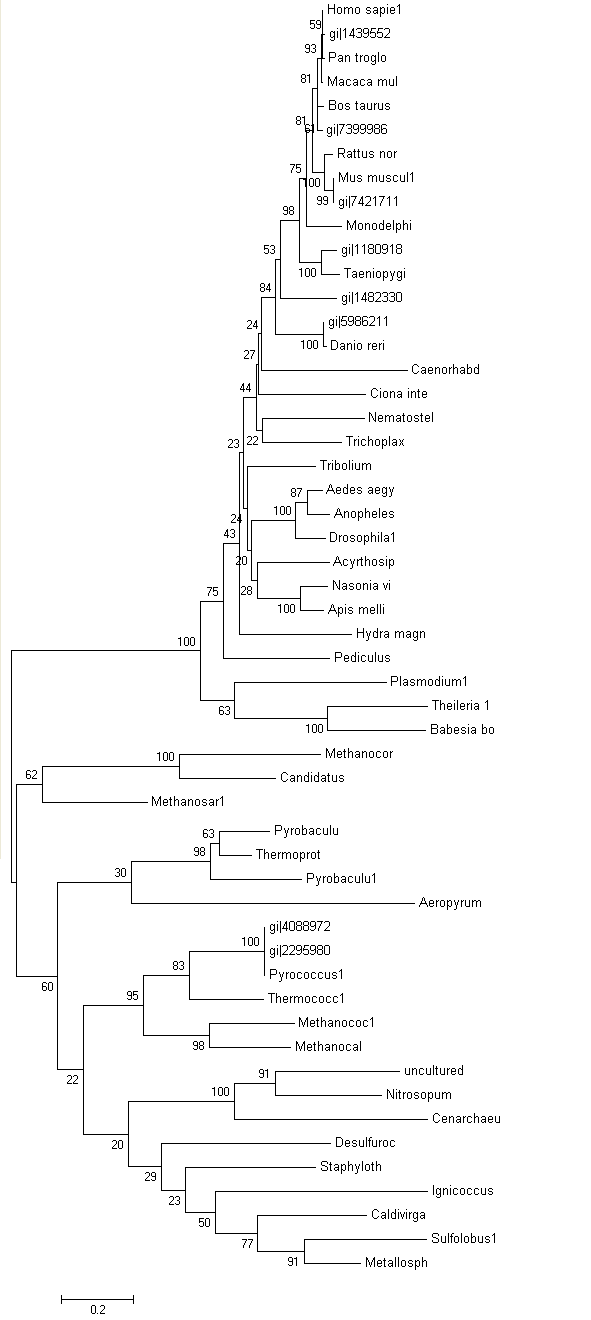
Phylogeny tree and Bootstrap
The phylogeny tree and boostrap revealed that PP-loop motif from ATP binding domain 4 and other related proteins are found in species of Archaea and Eukaryotes suggesting that this motif is highly conserved throughout evolution. However, Bacteria found to be lacking of this conserved sequences since none of the species belongs to Domain Bacteria. Nevertheless, based on STRING: functional protein association networks, some bacteria species still have protein sequences which belong to N-type ATP pyrophosphatase superfamily. BlastP are conducted to compare the protein sequences of ATPases from Pyrococcus furiosus and the bacteria species which under the clan of N-type ATP pyrophosphatase. The E value obtained from the result are below the E-value that is used as the cut-out-point. The bacteria species are Fusobacteruim nucleatum (1e-22), Caldicellulosiruptor saccharolyticus (1e-13),Campylobacter jejuni (4e-19),Chromobacterium violaceum (1e-21)and Polynucleobacter sp. (1e-24). These E value fall below the lowest E value based on Blastp from Pyrococcus furiosus and Macacca mulatta i.e: 5e-50 and 2e-25 respectively. These suggested that although some bacteria species still have protein sequences which belong to N-type ATP pyrophosphtase superfamily, they are distantly related to both archaea and eukarya. Based on the high protein sequence similarity between archaea and eukarya, it can be suggested that archeaa and eukarya are closely related compared to that for bacteria.
Bootstrap was conducted to test the reliability of the branching order of the phylogeny tree. Based on the bootstrap value, we can be confident with the order of the phylogeny tree thus allowing us to determine where speciation events occur. Bootstrap worked by performing 'pseudoreplicates' of multiple sequence alignments and in this project, 100 replicates was performed. The bootsrap tree can then be generated thus comparing the branching orders and distances of phylogeny tree. The bootstrap value for each branch are obtained in percentage which indicate the confidence of the branch being correct. If the value of bootstrap is less than 75%, the branching order is not very reliable and meaningless. If the value is between 90% and above, we can be confident that the branching orders are correct. Based on the bootstrap result, it was found that.....
Members of domain eukarya are nicely cluster at one side without any presence of other species from different domain i.e: Members from different domains are well-seperated. Therefore, it can be suggested that the evolutionary model for the PP-loop motif is hold since there is no evidence of the occurrence of lateral gene transfer.