ATP binding domain 4 Functions
My sequences:Putative n-type ATP pyrophosphatase from Pyrococcus furiosus, the Northeast Structural Genomics Target PfR23
AAHN-like superfamily
Adenine nucleotide alpha hydrolases, AANH_like (cl00292), superfamily includes members of N type ATP PPases, ATP sulphurylases Universal Stress Response protein and electron transfer flavoprotein (ETF). The domain forms a apha/beta/apha fold which binds to Adenosine nucleotide. Our sequence of interest is a putative n-type ATP pyrophosphatase that are under AAHN-like superfamily.
Pfam from Sanger suggests that N-type ATP Pyrophosphatase belongs to a family named ATP-binding 4 (PF01902), which contains a 200 amino acids long strongly conserved motif of SGGKD near the N-terminus.
The family ATP-binding 4 is a member of clan PP-loop (CL0039), which is comprised of 9 members: Arginossucinate synthase, Asn synthase, ATP binding 3, Exs B, NAD synthase, PAPS reductase, Thianmine biosynthesis protein, tRNA Methyl transferase and finally where our sequence belongs to -- ATP binding 4.
Although our protein sequence shows little homology with human ATP Binding domain 4 proteins, having % sequence ID = 36.8%, understanding the function of human ATPPD4 can help us infer functions of our sequence.
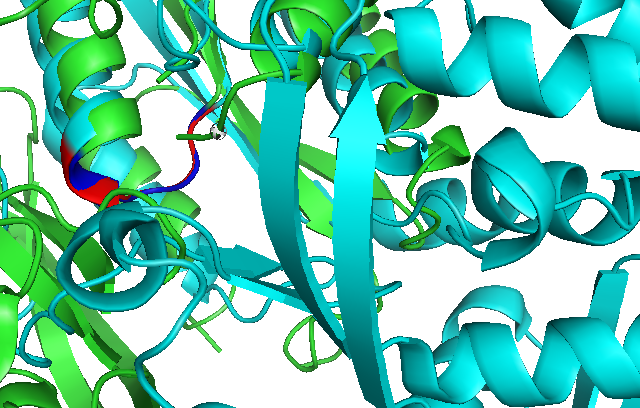
Dali analysis of Domain Alignment
Use of Dali reveals similar domain alignment of our sequence with Argininosuccinate Synthase (2nz2-A), which Z-score is 11.0% (indicatively significant domain similarity). Therefore, we use Pymol to align conserved residues of 1RU8 and 2NZ2 (align 1RU8 & i. 11-16, 2NZ2 & i. 11-15). Close structure resemblance is a strong indication of function resemblance.
Profunc Nest analysis for structural motifs
Nest analysis results for our sequence (chain A). Nests are structural motifs that are often found in functionally important regions of the protein structures. 7 nests have been located in our chain A.
High nest scores of nests 1-3 suggests that the first 3 sites are accessible to solvent, having significant conservative score compared to their parent residues or associated with those larger surface clefts. First 3 scores are all above 2.0 which are subjective of nests being relatively functionally significant.
Non-zero solvent accessibility: the percentage solvent accessible surface area of the residue's main chain nitrogen atom. As the first 3 residue ranges are non-zero then these atoms are accessible to solvent and capable of interacting with a binding anion, Leu4(A) in particular is very accessible to solvent.
Deeply clefted of nest 1 gives a strong indication of how deep in the cleft the NH atoms lie. As Ser12(A) and Gly14(A) lie particularly deep in the largest cleft 1, it is a strong indicator that they are functionally important.
Highly residue conserved in nests 1 and 2 has been determined from a multiple sequence alignment of the protein's sequence against BLAST hits from the UniProt sequence database. The conservation score of 1.0 in nests 1 and 2 indicates they are perfectly conserved.
1. Watson JD and Milner-White EJ (2002). A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi,psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol., 315, 171-82.
2. Watson JD and Milner-White EJ (2002). The conformations of polypeptide chains where the main-chain parts of successive residues are enantiomeric. Their occurrence in cation and anion-binding regions of proteins. J. Mol. Biol., 315, 183-191.
3. Pal D, Suhnel J and Weiss MS (2002). New principles of protein structure: nests, eggs - and what next? Angew. Chem. Int. Ed., 41, 4663-4665.
- ATP pyrophosphatase (ATP PPase) is used to assist lysidine formation using a lysine-specific loop and tRNA recognition domain
- Lysidine is a lysine-combined modified cytidine, locating at antcodon wobble position (34) of bacterial tRNA
- Usually ATP-pyrophosphatatse has a domain of GMP synthetase, for adding adenine and help lysine attack on Carbon atom.
http://www.pnas.org/content/102/21/7487.abstract
--
- NAD+ synthetase belongs to a member of the family of N-type ATP pyrophosphatase (ATP PPases)
- Some other members of N-type ATP pyrophosphatase include NAD+ synthetase, GMP synthetase, asparagine synthetase and argininosuccinate synthetase
- this family is characterised by strictly conserved fingerprint sequence Ser-Gly-Gly-X-Ser/ Thr-Ser/ Thr at P-loop (this is found by the comparison of 3-D structures of NAD+ synthetase and GMP synthetase
- Since they are in the same family, we can infer their structure similarities and propose the functions of our sequences. - Look for the ATP-binding sites on ref[11]
- Rizzi, M., et al., & Galizzi, A. (1996). Crystal structure of NH3-dependent NAD synthetase from Bacillus subtilis. EMBO J. 15, 5125-5134
--
- ([A/S]-[F/y]-S-G-G-[L/V]-D-T-[S/T] is a common consensus sequences that contains a glycine-rich motif that is common to a subset of ATP pyrophosphatases, which is known as 'N-type' ATP pyrophosphatases.
- N-type ATP pyrophosphatases all catalyses a substrate adenylation to activate a carbonyl C=O or carboxyl COO- group for the subsequent attack of a nitrogen nucleopile.
- This glycine-rich motif will form a modified P loop within the nucleotide binding domain, known as 'PP-motif'.
- Lemke, C, T. and Howell, P.L. 2001, The 1.6A Crystal Structure of E.coli Argininosuccinate Synthetase Suggests a Confomational Change during Catalysis, Structure 9:1153-1164.
--
Taxonomic distribution of Adenine nucleotide alpha hydrolases-like domains in all kingdoms.
Each node represents the features of a single taxonomic group, or organism. The nodes are arranged hierarchically in concentric rings. The parent taxon, located in the centre, leads recursively outwards towards its children. The size of the circle indicates the mean number of domains found per organism in a given taxonomic group. For individual organisms, it gives the actual number of domains.
Gough, J., Karplus, K., Hughey, R. and Chothia, C. (2001). "Assignment of Homology to Genome Sequences using a Library of Hidden Markov Models that Represent all Proteins of Known Structure." J. Mol. Biol., 313(4), 903-919.