ATP binding domain 4 Functions
Background
ATP pyrophosphatase activity commonly have a widespread motif of ([A/S]-[F/y]-S-G-G-[L/V]-D-T-[S/T] which is commonly conserved in many groups of enzymes including GMP synthetases, argininosuccinate synthetases and ATP sulfurylases. (Lemke et al. 2001) If this PP-motif also presents in our uncharacterised protein, this may give us a clue to its functions.
Consensus of alignment between our sequence and other ATP pyrophosphatase members demonstrates that this protein belongs to anabolic pathways and have ATP pyrophosphatase activity, meaning that they will participate in hydrolysing the Alpha-Beta bond in ATP. Moreover, the respective overall reaction schemes are different among different family members (see figure 1.0).
For examples, according to the table below, in the reaction scheme of argininosuccinate synthetase (Assy), the cleavage of ATP will provide energy for changing L-citrullin and L-Asp into L-argininosuccinate. Whereas, in ATP sulfurylase, ATP bond breaking will drive the sulfate transfer to AMP. Therefore, although our protein may involve in ATP high energy bond cleavage, we are not sure what type of subunits will our protein associate with the energy-requiring function since the reaction schemes of different proteins are different.
Structural-based methods:Matching folds by sequence comparison (SSM)
The ProFunc server is used to identify similarities in structure and hopefully can indicate conserved functions. The SSM (Secondary Structure Matching) program is used to compare the fold of our query protein (Putative n-type ATP pyrophosphatase) against the folds of the structural database in PDB. The secondary structure elements (SSEs) of two proteins are compared and ranked by Z-scores, which is computed from the matching value obtained from structural superpositions.
The fold match results for our query protein (1ru8 chain A) returns with 10 structural matches as shown in figure 2.0.
The top match (1ru8A) is our query sequence and so does the second match. The 3rd hit (2d13A) is a hypothetical dimeric protein from a related species, Pyrococcus horikoshii expressed from E. Coli. The result returns with a high value of 15 matched secondary structure elements. (as shown by No.SSE column) These two proteins have a high similarities in terms of helices and strands. This result is also coherent with Dali search too. However, the function of this protein is also unidentified and therefore of little use to us.
The hits beyond 3, colour-coded with orange and blue, are rather weak matches and unlikely hits; which can agrees with the Dali search in structural section(Krissinel & Henrick 2004).
Profunc Nest analysis for structural motifs
Nests are structural motifs that are particularly important for its functions. These nests are usually hidden deep within clefts to stay away from hydrophilic environment. These structural motifs are usually parts of hydrogen bond and may form charge-charge interactions with anionic groups or ligands. Nests are identified by computer programmes based on their alternative enantiomeric mainchain dihedral angles from the alpha and gamma regions of a Ramachandran plot. (Pal et la.2002) The results are shown as follows.
High nest scores of nests 1-3 in figure 3.0 suggests that the first 3 sites are accessible to solvent, having significant conservative score compared to their parent residues or associated with those larger surface clefts. First 3 scores are all above 2.0 which are subjective of nests being relatively functionally significant.
Non-zero solvent accessibility: the percentage solvent accessible surface area of the residue's main chain nitrogen atom. As the first 3 residue ranges are non-zero then these atoms are accessible to solvent and capable of interacting with a binding anion, Leu4(A) in particular is very accessible to solvent.
Deeply clefted of nest 1 gives a strong indication of how deep in the cleft the NH atoms lie (Watson & Milner-White 2002). As Ser12(A) and Gly14(A) lie particularly deep in the largest cleft 1, it is a strong indicator that they are functionally important. Highly residue conserved in nests 1 and 2 has been determined from a multiple sequence alignment of the protein's sequence against BLAST hits from the UniProt sequence database. The conservation score of 1.0 in nests 1 and 2 indicates they are perfectly conserved.
Cleft analysis
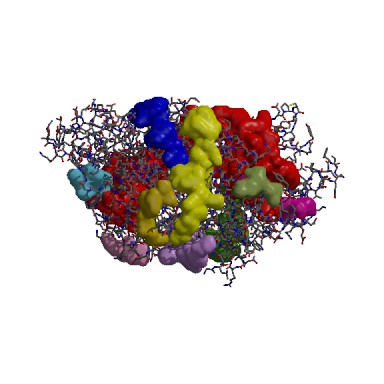
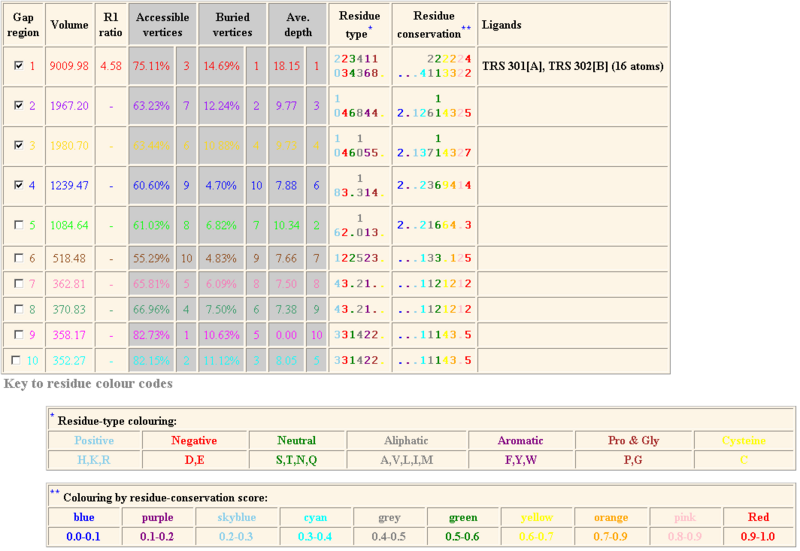
The following result table demonstrates the gaps in the order of decreasing volumes. Clefts and cavities in our protein have been colour-coded according to the table below. In gap region 1, this biggest cleft contains 42 residue conservations (shown in red letters in ‘Residue conservation’ column. A TRS which is a common crystallization solvent ligand was shown to bind to gap 1.
According to the cleft analysis provided in ProFunc in figure 4.2, the largest gap has a volume of 9009.98 in its dimeric form (Krissinel & Henrick 2004). The volume determined by ProFunc is more than twice larger than the value previously determined by CASTp (3385.7) in the structural section. It is suspected that the cleft will be expanded when it is dimerized form. Identification of the residue type may be helpful for us to identify how ATP binding domain 4 interact with substrate and deduce the reaction mechanism. The majority of the residues are aliphatic (43) and neutral (34). Fewer residues are negatively charged (23) and positively charged (20).
Adenine nucleotide alpha hydrolases, AANH_like (cl00292), superfamily includes members of N type ATP PPases, ATP sulphurylases Universal Stress Response protein and electron transfer flavoprotein (ETF). The domain forms a alpha/beta/alpha fold which binds to Adenosine nucleotide. Our sequence of interest is a putative n-type ATP pyrophosphatase that are under AAHN-like superfamily.
Pfam from Sanger suggests that N-type ATP Pyrophosphatase belongs to a family named ATP-binding 4 (PF01902), which contains a 200 amino acids long strongly conserved motif of SGGKD near the N-terminus.
The family ATP-binding 4 is a member of clan PP-loop (CL0039), which is comprised of 9 members (figure 5.0): Arginossucinate synthase, Asn synthase, ATP binding 3, Exs B, NAD synthase, PAPS reductase, Thianmine biosynthesis protein, tRNA Methyl transferase and finally where our sequence belongs to -- ATP binding 4.Therefore, we try to investigate on related family members to see which one is a potential candidate for functional modelling. From our structural studies (see structual section), results of Dali returns with two family members with greatest motif similarities, namely Argininosuccinate synthetase (AS) and Queuosine biosynthesis protein. However, only AS has shown close alignment with ATP binding domain 4 but not the other. The Dali alignment is shown in figure 6.0 below.
Dali analysis of Domain Alignment
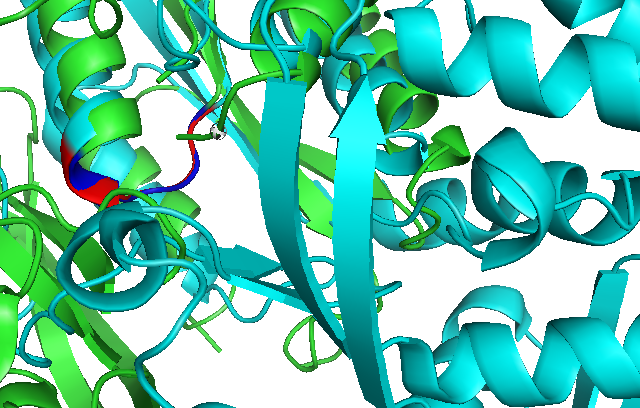
Use of Dali reveals similar domain alignment of our sequence with Argininosuccinate Synthase (2nz2-A), which Z-score is 11.0% (indicatively significant domain similarity). Therefore, we use Pymol to align conserved residues of 1RU8 and 2NZ2 (align 1RU8 & i. 11-16, 2NZ2 & i. 11-15). Close structure resemblance is a strong indication of function resemblance.
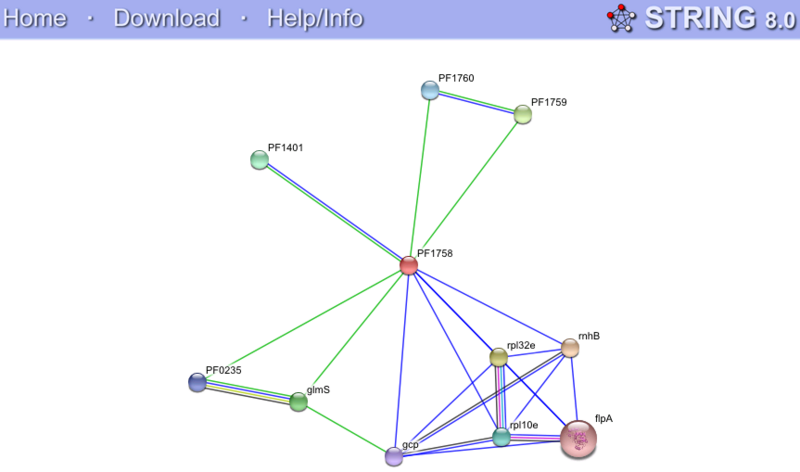
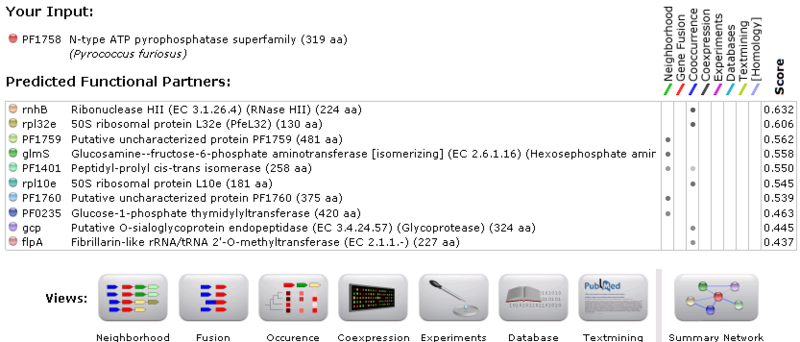
String Functional Partners Predictions
Green lines represent neighbourhood association. Blue lines represent coocurrence among different species. Weak association was shown in all hits. Our query protein (PF1758) as indicated by the red arrows is found immediate neighbourhood (within 300bp) with PF1760, PF1759 and glmS. This association indicates putative fusion events which means that functional related genes are usually inherited together. Unfortunately, PF1760 and PF1759 are putative uncharacterised proteins. Whereas, glmS is a glucosamine (fructose-6-phosphate aminotransferase), its association with our query protein is only discovered between one proteobacteria, meaning an insignificant association. Even though the result of String is of little use to us, it is suggested that N-type ATP pyrophosphatases present in both bacteria and archea. (Gough et la.2001) This finding assists us in studying the evolutionary history of proteins.