ATP binding domain 4 Structures: Difference between revisions
Anharmustafa (talk | contribs) |
Anharmustafa (talk | contribs) |
||
| Line 60: | Line 60: | ||
== Structure Similarities == | == Structure Similarities == | ||
[[Image:Dali33.PNG|framed|none]] | [[Image:Dali33.PNG|framed|none|'''Figure 6'''. The list of structural similarities to 1RU8 obtained using DALI]] | ||
''Z score'' , the statistical significance of the similarity between protein-of-interest and other neighbourhood proteins. The program optimizes a weighted sum of similarities of intramolecular distances. | ''Z score'' , the statistical significance of the similarity between protein-of-interest and other neighbourhood proteins. The program optimizes a weighted sum of similarities of intramolecular distances. | ||
| Line 71: | Line 71: | ||
''%id'', percentage of identical amino acids over structurally equivalent residues. | ''%id'', percentage of identical amino acids over structurally equivalent residues. | ||
== Structure Alignment == | == Structure Alignment == | ||
Revision as of 14:49, 1 June 2009
General information
General information from PDB indicates that :
(a) 1RU8 is a putative n-type ATP pyrophosphatase isolated from Pyrococcus furiosus, expressed in Escherichia Coli.
(b) Is a member of clan PP-loop
(c) Resolution of 2.7 angstroms, with an r-value of 0.218.
(d) Ligand chemical component identified as TRS (2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL).
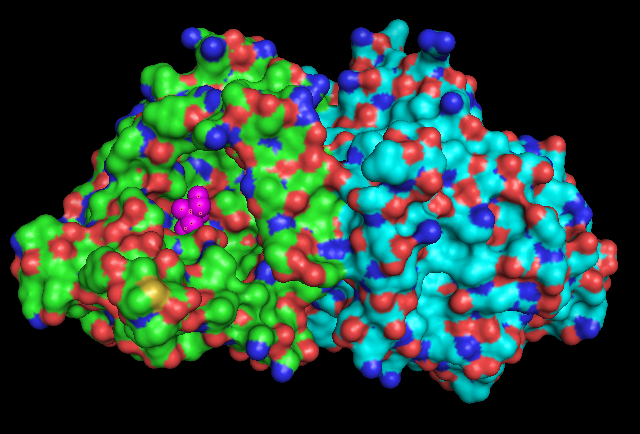
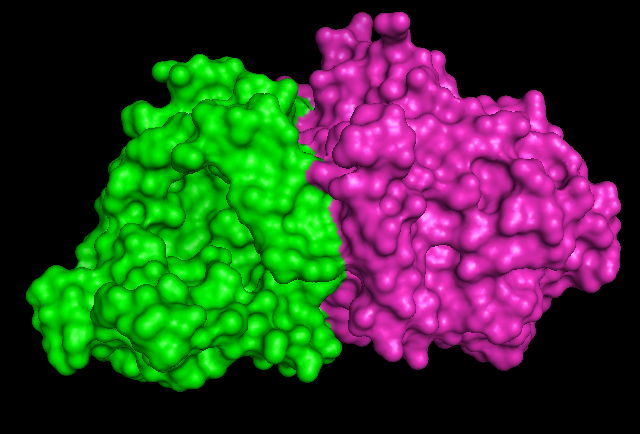
Surface Structure
Pymol Visualization
1RU8 is a dimer
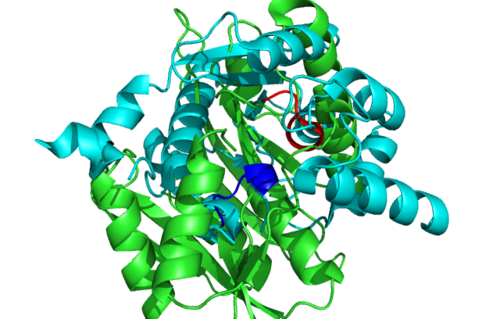
Secondary Structure and Location of P-loop (ATP binding site)
Electrostatic Surface Potential
Structure Similarities
Z score , the statistical significance of the similarity between protein-of-interest and other neighbourhood proteins. The program optimizes a weighted sum of similarities of intramolecular distances.
Root Mean Square Distance (RMSD), root-mean-square deviation of C-alpha atoms in the least-squares superimposition of the structurally equivalent C-alpha atoms. RMSD is not optimized and is only reported for information.
lali, the number of structurally equivalent residues.
nres, or the total number of amino acids in the hit protein.
%id, percentage of identical amino acids over structurally equivalent residues.
Structure Alignment
1RU8 and 2NZ2
1RU8 and 3BL5
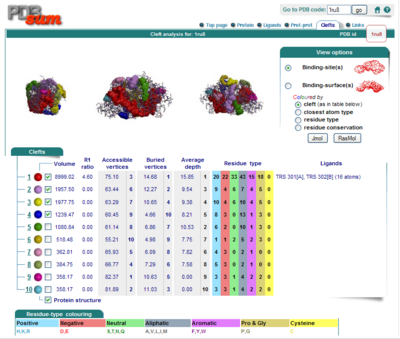
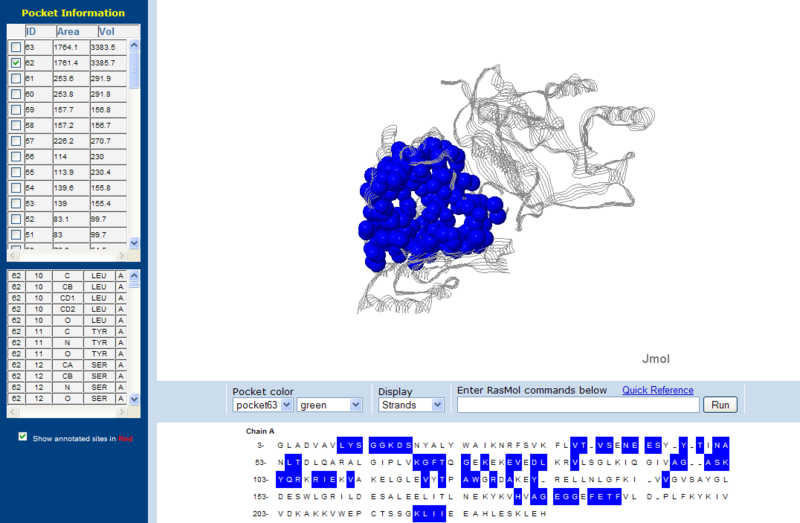
Surface Clefts
Surface Topography