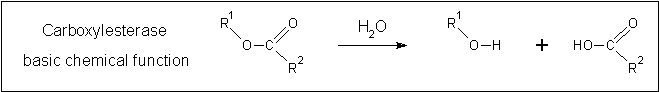Arylformamidase Function: Difference between revisions
Thomasparker (talk | contribs) No edit summary |
Thomasparker (talk | contribs) No edit summary |
||
| (14 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== Evidence from Similar Sequences == | |||
The putative annotation which was initially provided ''on the basis of...'' prompted a literature search using the term 'arylformamidase'. Pabarcus et al. (2007) have analysed the function of an arylformamidase present in the liver of ''Mus musculus''. | |||
Ironically, from the BLAST results, the most similar sequence to 2pbl for which there was functional information available was the arylformamidase characterised by Pabarcus et al. 2007. Using site-directed mutagenesis, a catalytic triad was identified - S162, D247 and H279. To elucidate any functional similarity between arylformamidase and 2pbl, conservation of the catalytic triad was assessed through a clustalW alignment (see figure ...). Both S162 and H279 were found to be conserved in relatively conserved regions of the alignment whereas D247 had undergone a semi-conserved substitution. These residues correlated to... The residues were located on the tertiary structure of 2pbl and determined to be sufficiently proximal to one another to permit catalysis (see figure...). | |||
[[Image:arylformamidase_alignment.png|centre|framed|'''Figure 3:''' ''Conservation of the catalytic triad between Arylformamidase and 2pbl.'']] | |||
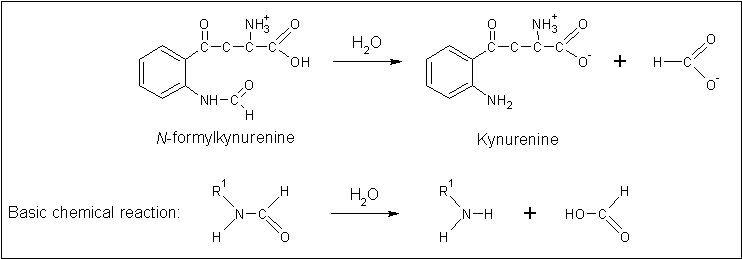
Arylformamidase forms part of the tryptophan degradation pathway... | |||
[[Image:Arylformadisae_reaction.gif|centre|framed|'''Figure 4:''' ''The reaction catalysed by Arylformamidase.'']] | |||
Carboxylesterases have a common reaction mechanism (see figure ...). This is somewhat similar to the arylformamidase reaction mechanism incorporating hydrolysis of a ... bond. | |||
[[Image:Carboxylesterase_reaction.gif|centre|framed|'''Figure 2:''' ''The fundamental reaction catalysed by carboxylesterases.'']] | |||
Evolutionary... | |||
[[Arylformamidase | Return to the main page...]] | [[Arylformamidase | Return to the main page...]] | ||
Latest revision as of 11:26, 3 June 2008
Evidence from Similar Sequences
The putative annotation which was initially provided on the basis of... prompted a literature search using the term 'arylformamidase'. Pabarcus et al. (2007) have analysed the function of an arylformamidase present in the liver of Mus musculus. Ironically, from the BLAST results, the most similar sequence to 2pbl for which there was functional information available was the arylformamidase characterised by Pabarcus et al. 2007. Using site-directed mutagenesis, a catalytic triad was identified - S162, D247 and H279. To elucidate any functional similarity between arylformamidase and 2pbl, conservation of the catalytic triad was assessed through a clustalW alignment (see figure ...). Both S162 and H279 were found to be conserved in relatively conserved regions of the alignment whereas D247 had undergone a semi-conserved substitution. These residues correlated to... The residues were located on the tertiary structure of 2pbl and determined to be sufficiently proximal to one another to permit catalysis (see figure...).
Arylformamidase forms part of the tryptophan degradation pathway...
Carboxylesterases have a common reaction mechanism (see figure ...). This is somewhat similar to the arylformamidase reaction mechanism incorporating hydrolysis of a ... bond.
Evolutionary...