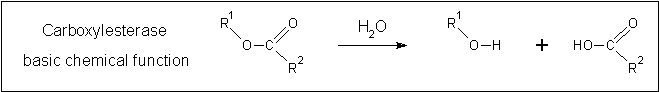Arylformamidase Function: Difference between revisions
From MDWiki
Jump to navigationJump to search
Thomasparker (talk | contribs) No edit summary |
Thomasparker (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
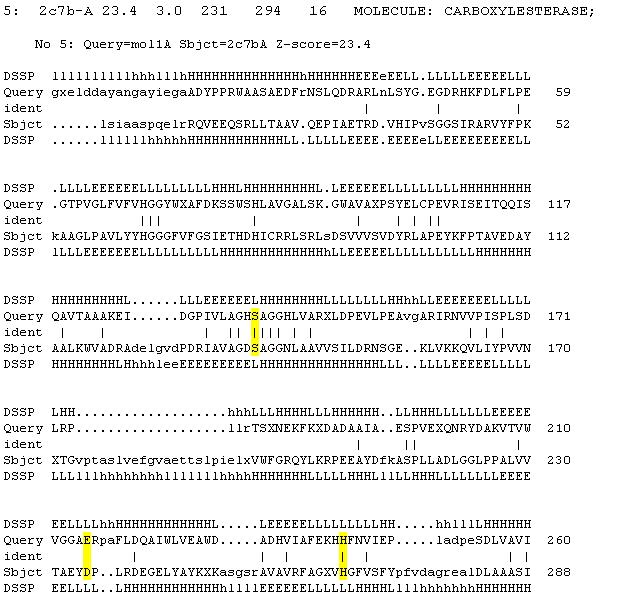
From the DALI output, 2c7b was found to be the most similar structure to 2pbl having a Z-score of 23.4, 16% sequence identity and a root mean square deviation of 3.0. Conservation of the catalytic triad of 2cb7 as identified by ... was analysed for 2pbl (see figure 1). | From the DALI output, 2c7b was found to be the most similar structure to 2pbl having a Z-score of 23.4, 16% sequence identity and a root mean square deviation of 3.0. Conservation of the catalytic triad of 2cb7 as identified by ... was analysed for 2pbl (see figure 1). | ||
[[Image:2cb7alignment.jpg | [[Image:2cb7alignment.jpg|none|framed|'''Figure 1:''' ''Conservation of the catalytic triad between 2cb7 and 2pbl.'']] | ||
A review of the DALI output revealed many structures similar to 2pbl with little sequence identity. The most similar of these, 2cb7, is a thermophilic and thermostable carboxylesterase. The catalytic triad responsible for its activity has been identified, and in determining whether 2pbl shared similar functionality, conservation of the catalytic triad between the two proteins was assessed (see figure 1). | A review of the DALI output revealed many structures similar to 2pbl with little sequence identity. The most similar of these, 2cb7, is a thermophilic and thermostable carboxylesterase. The catalytic triad responsible for its activity has been identified, and in determining whether 2pbl shared similar functionality, conservation of the catalytic triad between the two proteins was assessed (see figure 1). | ||
[[Image:Carboxylesterase_reaction.gif | [[Image:Carboxylesterase_reaction.gif|none|framed|'''Figure 2:''' ''The fundamental reaction catalysed by carboxylesterases.'']] | ||
== Evidence from Similar Sequences == | == Evidence from Similar Sequences == | ||
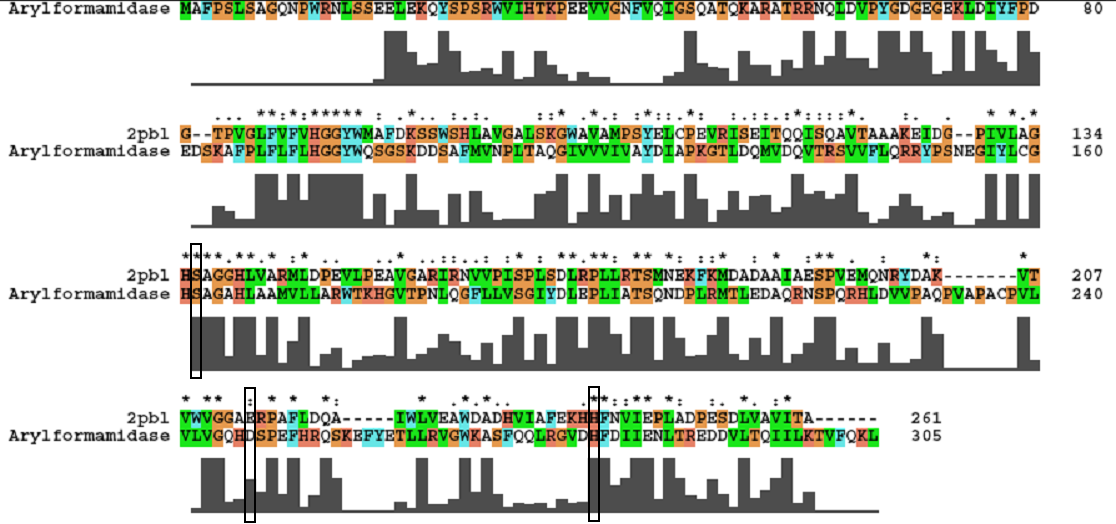
[[Image:arylformamidase_alignment.jpg | [[Image:arylformamidase_alignment.jpg|none|framed|'''Figure 3:''' ''Conservation of the catalytic triad between Arylformamidase and 2pbl.'']] | ||
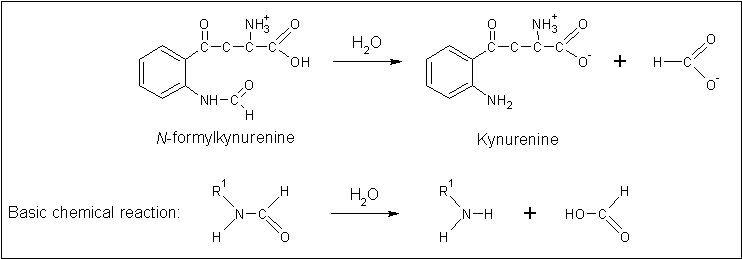
[[Image:Arylformadisae_reaction.gif | [[Image:Arylformadisae_reaction.gif|none|framed|'''Figure 4:''' ''The reaction catalysed by Arylformamidase.'']] | ||
[[Arylformamidase | Return to the main page...]] | [[Arylformamidase | Return to the main page...]] | ||
Revision as of 06:03, 31 May 2008
From the DALI output, 2c7b was found to be the most similar structure to 2pbl having a Z-score of 23.4, 16% sequence identity and a root mean square deviation of 3.0. Conservation of the catalytic triad of 2cb7 as identified by ... was analysed for 2pbl (see figure 1).
A review of the DALI output revealed many structures similar to 2pbl with little sequence identity. The most similar of these, 2cb7, is a thermophilic and thermostable carboxylesterase. The catalytic triad responsible for its activity has been identified, and in determining whether 2pbl shared similar functionality, conservation of the catalytic triad between the two proteins was assessed (see figure 1).