Arylformamidase Function Slide 5: Difference between revisions
From MDWiki
Jump to navigationJump to search
Thomasparker (talk | contribs) No edit summary |
Thomasparker (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
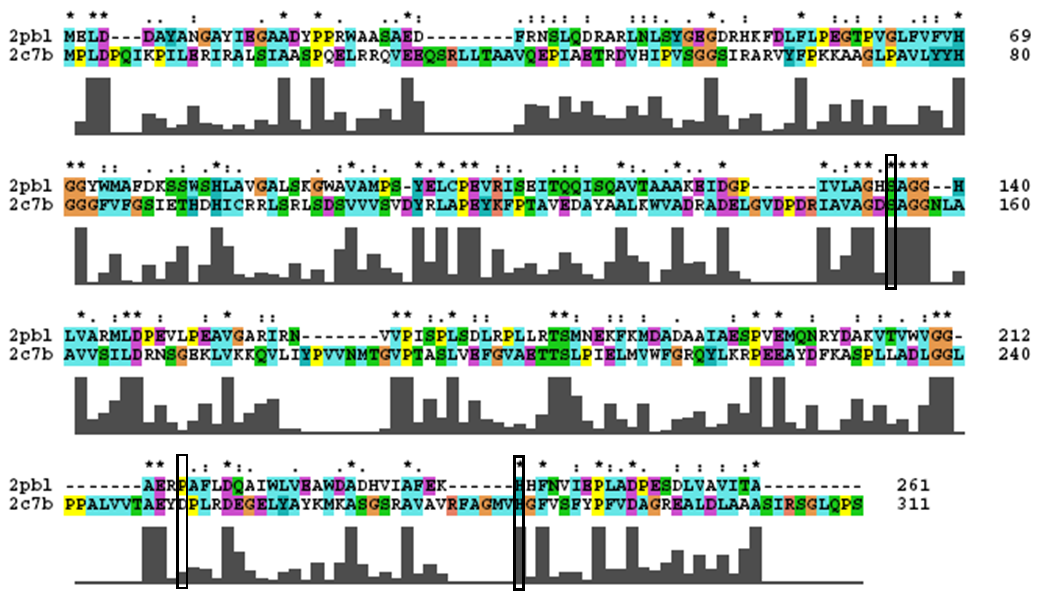
== | == Evidence from Similar Structures == | ||
- Catalytic triad identified in paper - Ser154, Asp251, and His281. | |||
- To assess functional similarity, conservation of the catalytic triad was analysed. | |||
''' | [[Image:2c7b_alignment.png|centre|framed|'''Conservation of the catalytic triad between 2cb7 and 2pbl.''']] | ||
- Catalytic triad conserved. | |||
- Note: difference in clustalW alignment from DALI sequence alignment. Unstable region of alignment? | |||
[[Arylformamidase Function Slide 4| ...Previous slide ]]|[[Arylformamidase| Return to the main page ]]|[[Arylformamidase Function Slide 6| Next slide... ]] | [[Arylformamidase Function Slide 4| ...Previous slide ]]|[[Arylformamidase| Return to the main page ]]|[[Arylformamidase Function Slide 6| Next slide... ]] | ||
Revision as of 12:30, 8 June 2008
Evidence from Similar Structures
- Catalytic triad identified in paper - Ser154, Asp251, and His281.
- To assess functional similarity, conservation of the catalytic triad was analysed.
- Catalytic triad conserved.
- Note: difference in clustalW alignment from DALI sequence alignment. Unstable region of alignment?