Arylformamidase Sequence & Homology: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
[[Image:bacterTREE.jpg]] | [[Image:bacterTREE.jpg]] | ||
== Method == | |||
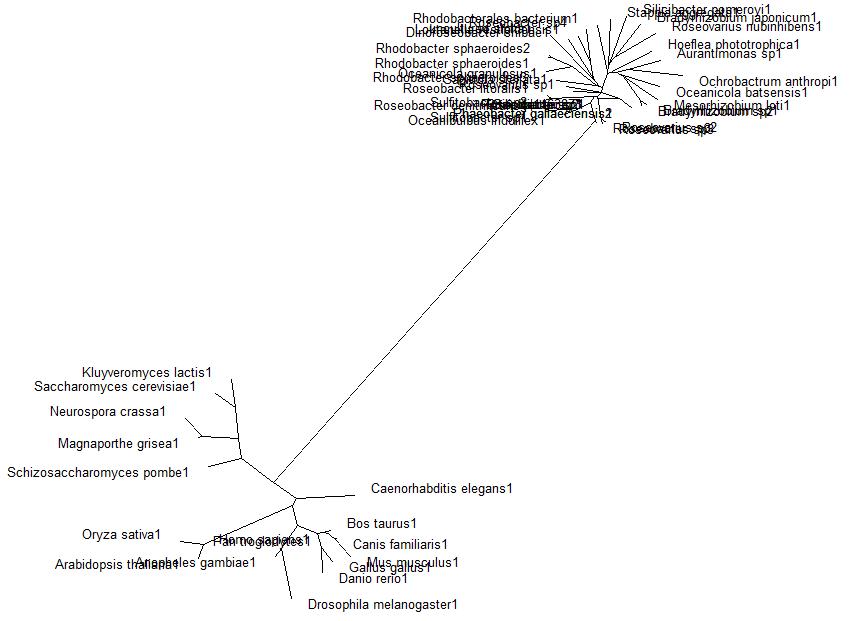
A BLAST search was performed on the bacterial sequence of Arylformamidase. The top scoring matches to an E-value of 3e-054, 35 sequences in total, were selected. Eukaryotic homologous sequences sequences found using HOMOLOGENE. These included vertebrates, invertebrates, plants and single-celled eukaryotes. These were appended to the list and a multple sequece alignment was performed using CLUSTAL X. The alignment revealed several conserved regions accross all species, thereby indicating a high level of conservation from Bacteria through Eukaryota. | |||
Revision as of 05:32, 27 May 2008
Unrooted phylogenetic tree ofhgest scoring BLAST search of bacteria sequnces and homologous eukaryotic sequences sourced from HOMOLOGENE:
Method
A BLAST search was performed on the bacterial sequence of Arylformamidase. The top scoring matches to an E-value of 3e-054, 35 sequences in total, were selected. Eukaryotic homologous sequences sequences found using HOMOLOGENE. These included vertebrates, invertebrates, plants and single-celled eukaryotes. These were appended to the list and a multple sequece alignment was performed using CLUSTAL X. The alignment revealed several conserved regions accross all species, thereby indicating a high level of conservation from Bacteria through Eukaryota.