Arylformamidase Sequence & Homology: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
Using the query sequence pdb:2pbl "Arylformamidase", a BLAST search was performed using a non-redundant database. The top scoring matches to an E-value of 3e-054, 35 sequences in total, were selected. Eukaryotic homologous sequences sequences were found using HOMOLOGENE. These were appended to the list and a multple sequece alignment was performed using CLUSTAL X. | Using the query sequence Target 13, pdb:2pbl "Arylformamidase", a BLAST search was performed using a non-redundant database. The top scoring matches to an E-value of 3e-054, 35 sequences in total, were selected. Eukaryotic homologous sequences sequences were found using HOMOLOGENE. These were appended to the list and a multple sequece alignment was performed using CLUSTAL X. | ||
The data output from the multiple sequence alignment was bootstrapped 100 times and a phylogenetic tree was created using the neighbour-joining method. The program FigTree was used to create the visual representation of this tree(Figure 1). | The data output from the multiple sequence alignment was bootstrapped 100 times and a phylogenetic tree was created using the neighbour-joining method. The program FigTree was used to create the visual representation of this tree(Figure 1). | ||
| Line 18: | Line 18: | ||
The alignment revealed several conserved regions accross all species, thereby indicating a high level of conservation from Bacteria through Eukaryota. These included vertebrates, invertebrates, plants and single-celled eukaryotes. | The alignment revealed several conserved regions accross all species, thereby indicating a high level of conservation from Bacteria through Eukaryota. These included vertebrates, invertebrates, plants and single-celled eukaryotes. | ||
The query sequence "Arylformamidase" grouped with bacterial sequences, shown cloured in Red and Purple. The bootstrap values reveal low confidence with many of the nodes occurring lower down on the phylogenetic tree revealing a possible explanation for certain closely related species to be grouped into separate clades. However, despite low bootstrap scores, the grouping does reliably separate prokaryotes from eukaryotes and the eukaryotes themsselves are clearly distinguished between yeasts and moulds (shown in Blue), plants (Green), invertebrates (Orange) and vertebrates (shown in Black). | |||
Revision as of 08:07, 28 May 2008
Method
Using the query sequence Target 13, pdb:2pbl "Arylformamidase", a BLAST search was performed using a non-redundant database. The top scoring matches to an E-value of 3e-054, 35 sequences in total, were selected. Eukaryotic homologous sequences sequences were found using HOMOLOGENE. These were appended to the list and a multple sequece alignment was performed using CLUSTAL X.
The data output from the multiple sequence alignment was bootstrapped 100 times and a phylogenetic tree was created using the neighbour-joining method. The program FigTree was used to create the visual representation of this tree(Figure 1).
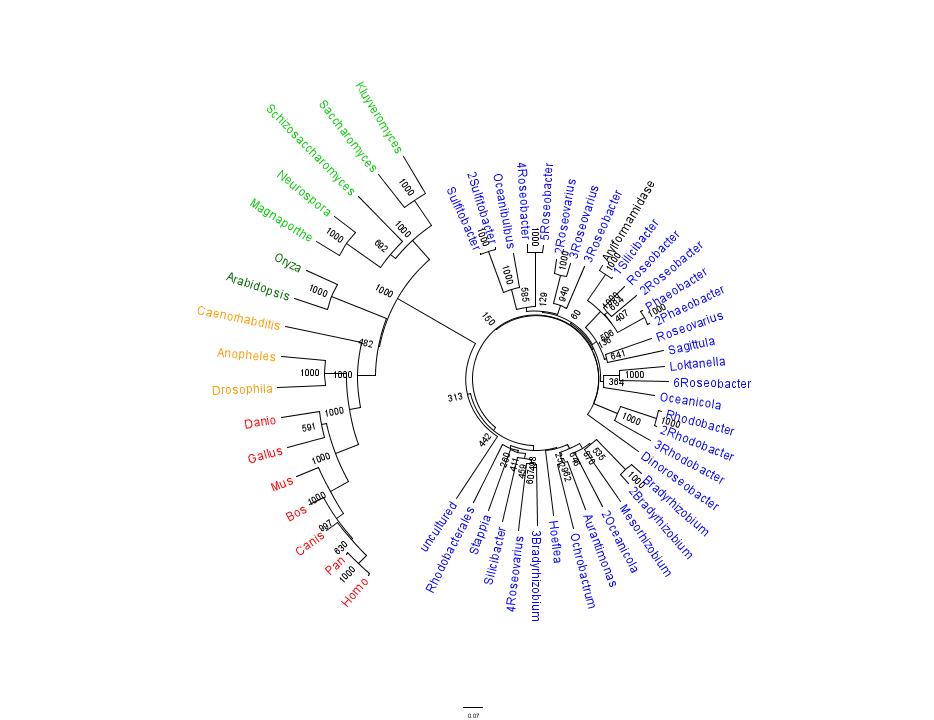
Figure 1.
Unrooted phylogenetic tree of highest scoring results from a BLAST search of bacterial sequnces using a non-redundant database and homologous eukaryotic sequences sourced from HOMOLOGENE:
Results
The alignment revealed several conserved regions accross all species, thereby indicating a high level of conservation from Bacteria through Eukaryota. These included vertebrates, invertebrates, plants and single-celled eukaryotes.
The query sequence "Arylformamidase" grouped with bacterial sequences, shown cloured in Red and Purple. The bootstrap values reveal low confidence with many of the nodes occurring lower down on the phylogenetic tree revealing a possible explanation for certain closely related species to be grouped into separate clades. However, despite low bootstrap scores, the grouping does reliably separate prokaryotes from eukaryotes and the eukaryotes themsselves are clearly distinguished between yeasts and moulds (shown in Blue), plants (Green), invertebrates (Orange) and vertebrates (shown in Black).