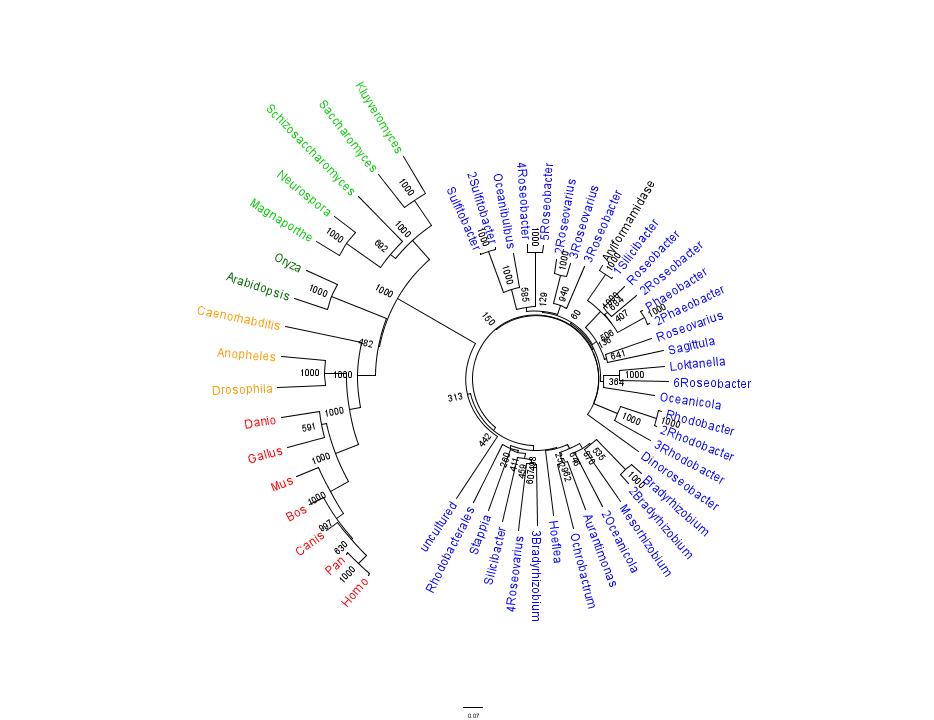
Arylformamidase Sequence & Homology
Method
A BLAST search was performed on the bacterial sequence of Arylformamidase. The top scoring matches to an E-value of 3e-054, 35 sequences in total, were selected. Eukaryotic homologous sequences sequences were found using HOMOLOGENE. These included vertebrates, invertebrates, plants and single-celled eukaryotes. These were appended to the list and a multple sequece alignment was performed using CLUSTAL X. The alignment revealed several conserved regions accross all species, thereby indicating a high level of conservation from Bacteria through Eukaryota.
The data output from the multiple sequence alignment was bootstrapped 100 times and a phylogenetic tree was created using the neighbour-joining method. The program FigTree was used to create the visual representation of this tree.
This gave the following tree:
Unrooted phylogenetic tree of highest scoring results from a BLAST search of bacterial sequnces using a non-redundant database and homologous eukaryotic sequences sourced from HOMOLOGENE: