Arylformamidase Structure: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
''' | == 1. Structure of Arylformamidase == | ||
'''Arylformamidase''' | |||
[[Image:Whole protein.png]] | |||
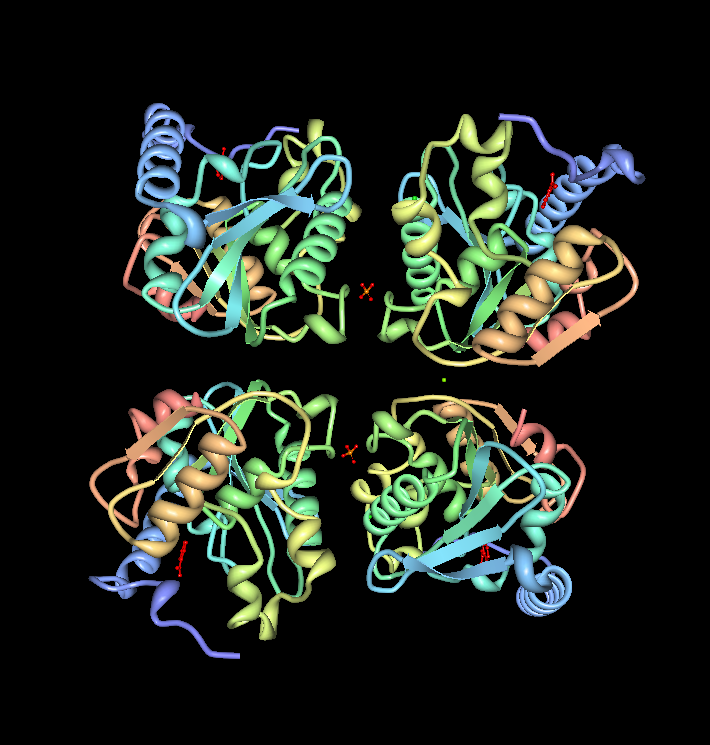
''The image above shows the chains A (upper right), B (upper left), C (lower right) & D (lower left) interacting. The molecules in the middle of chains A & B and chains C & D is phosphate ion (PO4). The green molecule between chain B & D is a magnesium ion (Mg).'' | |||
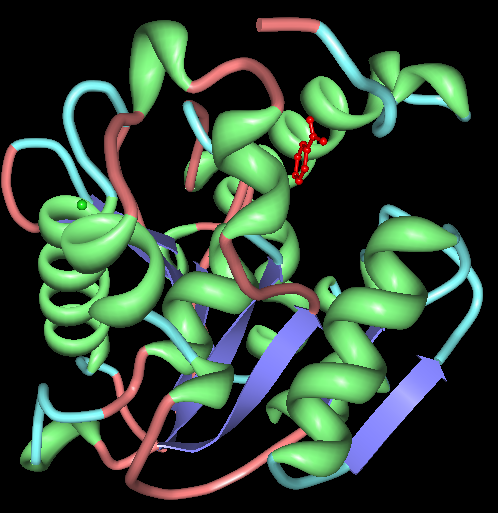
'''''Chain A'' of arylformamidase''' | |||
[[Image:chain_A.jpg]] | [[Image:chain_A.jpg]] | ||
| Line 5: | Line 21: | ||
''The red molecule in the middle is an unknown ligand containing a ring composed of 9 oxygen molecules. The green sphere is a chloride ion.'' | ''The red molecule in the middle is an unknown ligand containing a ring composed of 9 oxygen molecules. The green sphere is a chloride ion.'' | ||
| Line 16: | Line 29: | ||
=='''2. Putative Thioesterase'''== | |||
Thioesterases split ester groups into acid and alcohol in the presence of water, specifically at a thiol group | |||
Thioesterases or thiolester hydrolases are members of E.C.3.1.2. | Thioesterases or thiolester hydrolases are members of E.C.3.1.2. | ||
Our structure resembles carboxylesterases (hydrolases) hence it is predicted that it has similar cellular activity. | |||
| Line 36: | Line 53: | ||
''' | |||
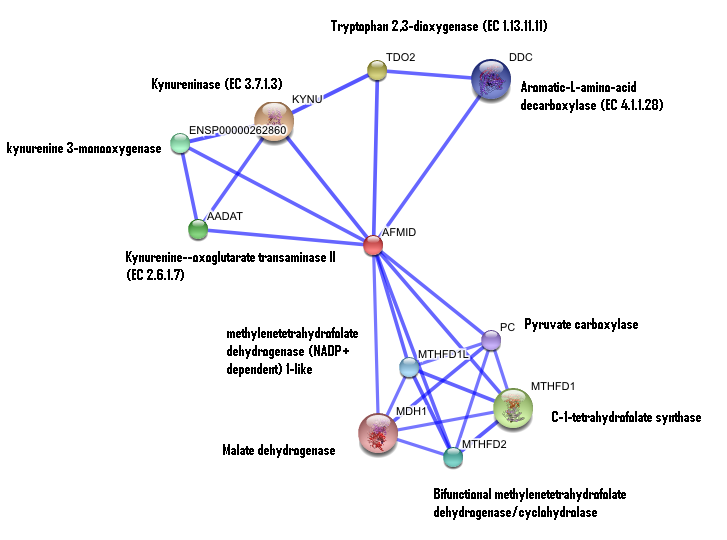
== '''3. Interaction of human arylformamidase with other proteins''' == | |||
[[Image:Confidence_interaction_with_names.png]] | [[Image:Confidence_interaction_with_names.png]] | ||
This image was generated from ["STRINGS"] | |||
== '''4. DALI OUTPUT''' == | |||
[[Image:DALI RESULT.txt]] | [[Image:DALI RESULT.txt]] | ||
| Line 73: | Line 97: | ||
== '''5. Secondary structure analysis''' == | |||
'''PDBSum output for arylformamidase''' | '''PDBSum output for arylformamidase''' | ||
| Line 84: | Line 107: | ||
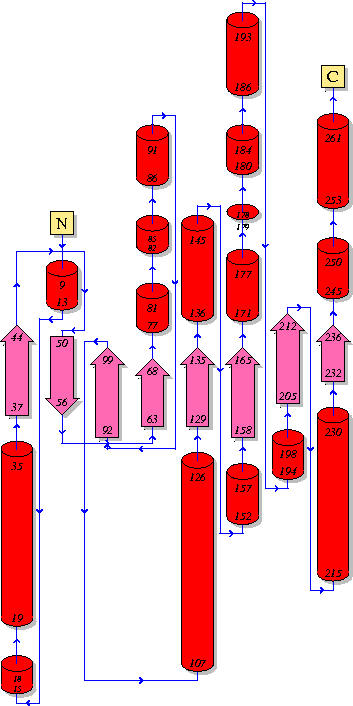
'''Topology''' | == '''6. Topology''' == | ||
[[Image:Topology.gif]] | [[Image:Topology.gif]] | ||
[[Arylformamidase | Return to the main page...]] | [[Arylformamidase | Return to the main page...]] | ||
Revision as of 02:37, 17 May 2008
1. Structure of Arylformamidase
Arylformamidase
The image above shows the chains A (upper right), B (upper left), C (lower right) & D (lower left) interacting. The molecules in the middle of chains A & B and chains C & D is phosphate ion (PO4). The green molecule between chain B & D is a magnesium ion (Mg).
Chain A of arylformamidase
The red molecule in the middle is an unknown ligand containing a ring composed of 9 oxygen molecules. The green sphere is a chloride ion.
2. Putative Thioesterase
Thioesterases split ester groups into acid and alcohol in the presence of water, specifically at a thiol group
Thioesterases or thiolester hydrolases are members of E.C.3.1.2.
Our structure resembles carboxylesterases (hydrolases) hence it is predicted that it has similar cellular activity.
3. Interaction of human arylformamidase with other proteins
This image was generated from ["STRINGS"]
4. DALI OUTPUT
Carboxylesterase A chain
PDB link title
Archeaon Carboxylesterase (Chains A B C D)
File:Carboxylesterase (archaeon).txt
PDB link title
5. Secondary structure analysis
PDBSum output for arylformamidase
PDBSUM [1]