Arylformamidase Structure
Methods
The structure of arylfromamidase was obtained from RCSB Protein Data Bank (PDB ID: 2PBL). http://www.rcsb.org/pdb/home/home.do
The predicted interaction arylformamidase with other proteins was determined using the STRING database (STRING: Search Tool for the Retrieval of Interacting Genes/Proteins). http://string.embl.de//
The DALI database was used for the structural comparison of arylformamidase with other proteins. http://ekhidna.biocenter.helsinki.fi/dali_server/
PDBsum database was used to determine the secondary structure of arylformamidase. http://www.ebi.ac.uk/pdbsum/
Results
Structure of Arylformamidase
Arylformamidase (All Chains)
The image above shows the chains A (upper right), B (upper left), C (lower right) & D (lower left) interacting. The molecules in the middle of chains A & B and chains C & D is phosphate ion (PO4). The green molecule between chain B & D is a magnesium ion (Mg). These ions aren't biologically significant and could only be an artefact. Those chains exist as indivitual functional units.
Chain A of arylformamidase
The red molecule in the middle is an unknown ligand containing a ring composed of 9 oxygen molecules. The green sphere is a chloride ion.
The protein backbone is coloured by conformation type:
Turn - blue
Coil- pink
Helix- green
strand- purple
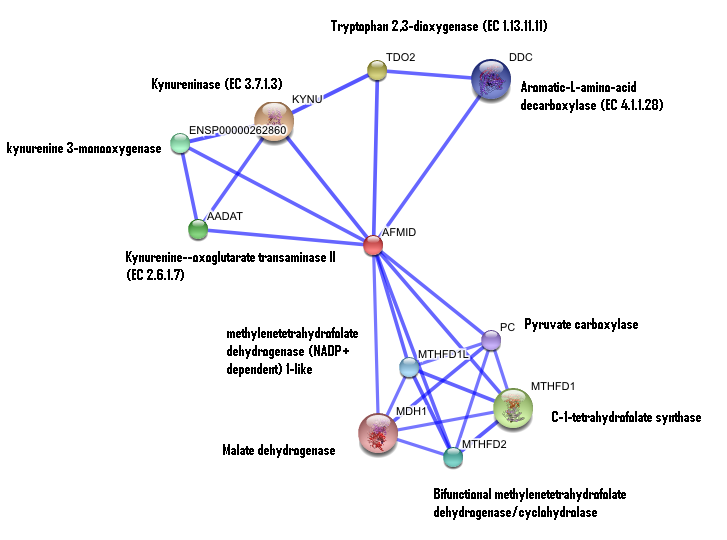
Interaction of human arylformamidase (AFMID) with other proteins
The interaction between the proteins have been determined from curated STRING database (significant score). However there is no significant evidence for:
1- Neighborhood in the genome
2- Gene fusions
3- Cooccurence across genomes
4- Co-Expression
5- Experimental/Biochemical data
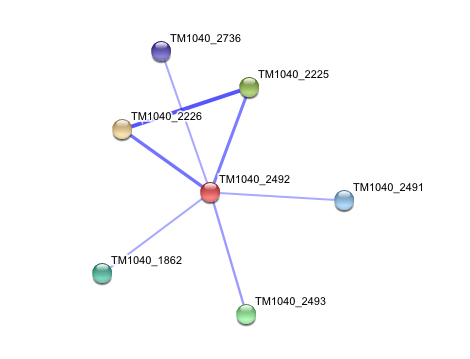
Interaction of Silicibacter Sp. arylformamidase (AFMID) with other proteins
TM1040_2226 Tryptophan 2,3-dioxygenase (279 aa)
TM1040_2225 Kynureninase (396 aa)
TM1040_2493 Succinic semialdehyde dehydrogenase (490 aa)
TM1040_1862 Hypothetical protein (212 aa)
TM1040_2491 Creatinase (402 aa)
TM1040_2736 Transketolase, putative (794 aa)
There is no significant evidence for these interactions (score= ~0.5)
DALI OUTPUT
The DALI tool produces proteins that are structurally similar to the protein of interest.
The search result showed similarities to mostly carboxylesterases/hydrolases. Hence there is strong evidence that our protein might also be a carboxylesterase.
Metagenomic Archea Carboxylesterase (Chain A ONLY)
PDB link title
Note: Chain B not shown
Archaeoglobus fulgidus Carboxylesterase (Chain A ONLY)
File:Carboxylesterase (archaeon).txt
PDB link title
Note: Chains B, C & D not shown
Both of the above Archaeal carboxylesterases' chains exist as monomers (from literature). Hence it is expected that our protein exists as a monomer but during crystalization it interacts with its chains.
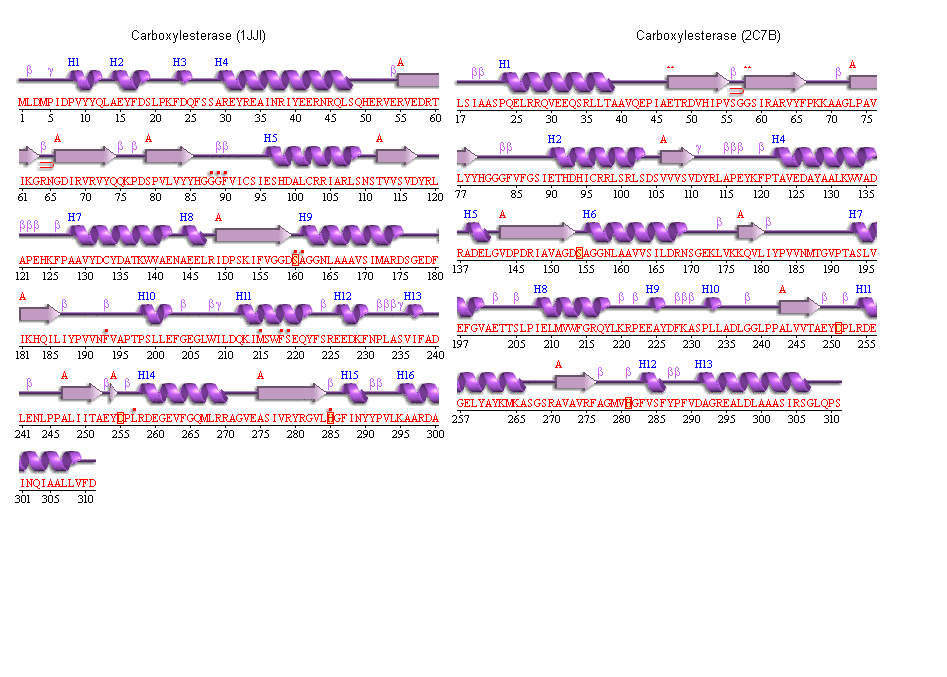
Secondary structure analysis
PDBSum output for arylformamidase
PDBSUM [1]
Archeon Carboxylesterase secondary structure
The conservation of the ser/his/asp catalytic triad
Yellow indicates conservation
Blue indicates semi-conservation
The above image shows the conserved residues of catalytic triad in arylformamidase, with the unknown ligand (Blue) protruding from a surface groove.
SER 136
HIS 241
GLU 214
The below image shows the distance between the catalytic triad conserved residues and how each amino acid is linked to a turn region.
The conserved catalytic triad in Metagenomic Archea Carboxylesterase (PDB ID 2C7B)