COASY results
Appearance of Coenzyme A Synthase
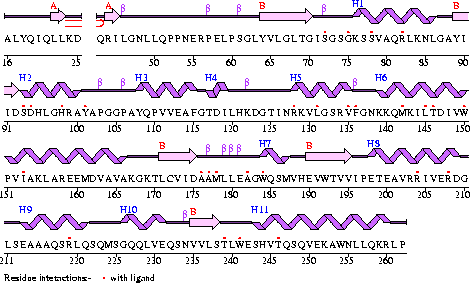
Conezyme A Synthase is structurally composed of seven strands, eleven helices and thirteen beta turns (EMBL EBI, 2005) (see Figure 1). Analysis of structurally related proteins (Holm & Sander, 1993) showed a trend for transferase (RCSB, 2007) class proteins with Rossmann class folds (Rossmann, 1973). The Rossmann topologies of fold’s are an alpha-beta class fold that forms a three or more layer beta strand sandwich alternating with alpha helices (beta-alpha-beta-alpha-beta). PFAM classification placed these into either the Cytidylyltransferase or Dephospho-CoA kinase familes, matching the two domains of Coenzyme A Synthase. Those classified under the Dephospho-CoA kinase type were commonly of the P-loop containing nucleotide triphosphate hydrolases (Sanger Institute, 2005) homology whilst those classified as Cytidylyltransferase were of the Tyrosol-Transfer RNA Synthetase (Sanger Institute, 2005). As the majority of the structurally related proteins identified contained the DPCK domain solely, and the motif for a P-loop was identified in the Conzyme A Synthase sequence (Bairoch, Bucher, & Hofmann, 1997) it is suggested that Coenzyme A Synthase is also of the P-loop containing nucleotide triphosphate hydrolases homology of folds.
Figure 1
The secondary structure of Mus musculus with indicated ligand interaction sites (EMBL EBI, 2005).
Figure 2
Structural alignment of structurally related proteins to Mus. musculus Coenzyme A Synthase