Chromosome 1 open reading frame 41 Evolution: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 23: | Line 23: | ||
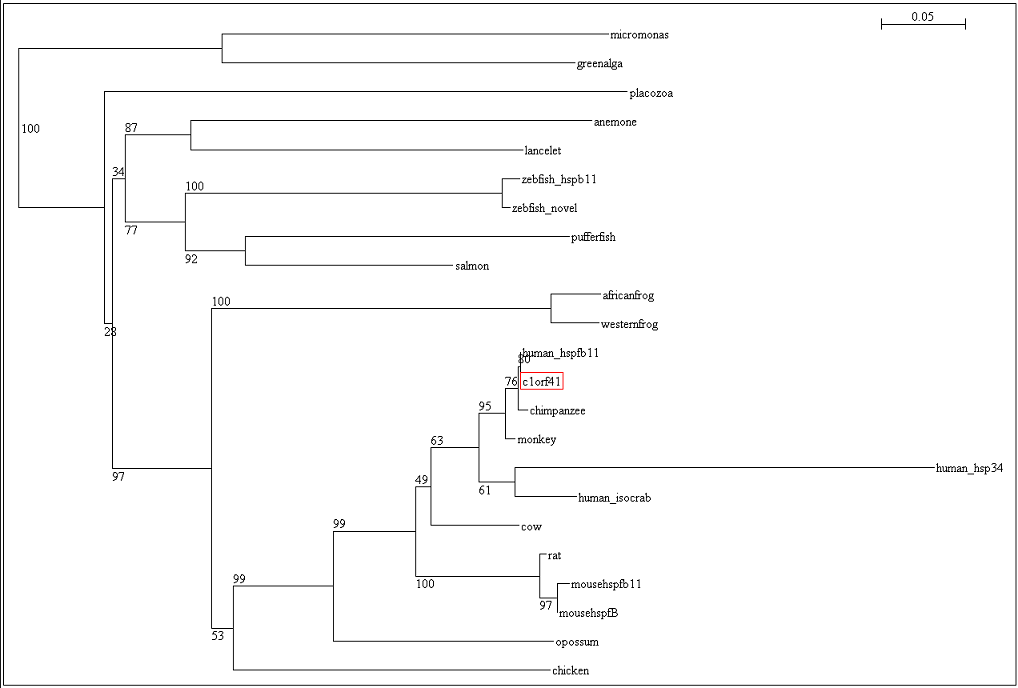
After obtaining the multiple sequence alignment, phylogenetic tree was construct based on the result from multiple sequence alignment. Bootstrap values are shown on each branch. | After obtaining the multiple sequence alignment, phylogenetic tree was construct based on the result from multiple sequence alignment. Bootstrap values are shown on each branch. | ||
[[Image:Tree. | [[Image:Tree.png]] | ||
Revision as of 08:45, 9 June 2009
Blast
The above shows the result from BLASTP search. The sequences of these proteins are aligned to observe the conserved regions.
ClustalX
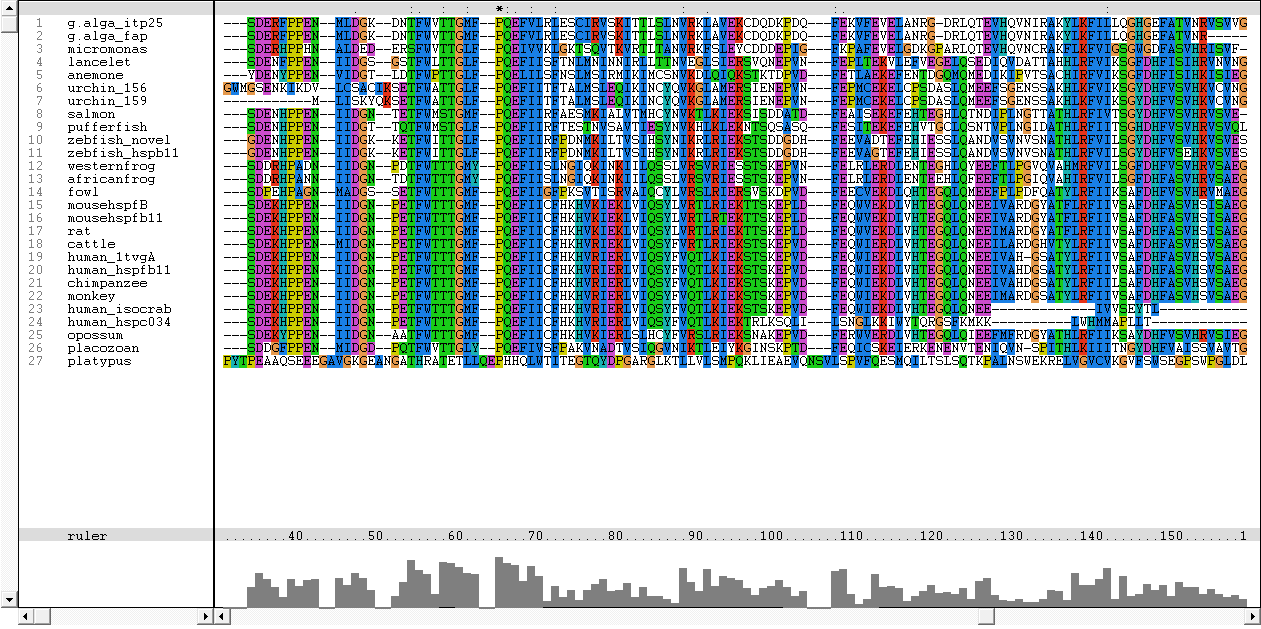
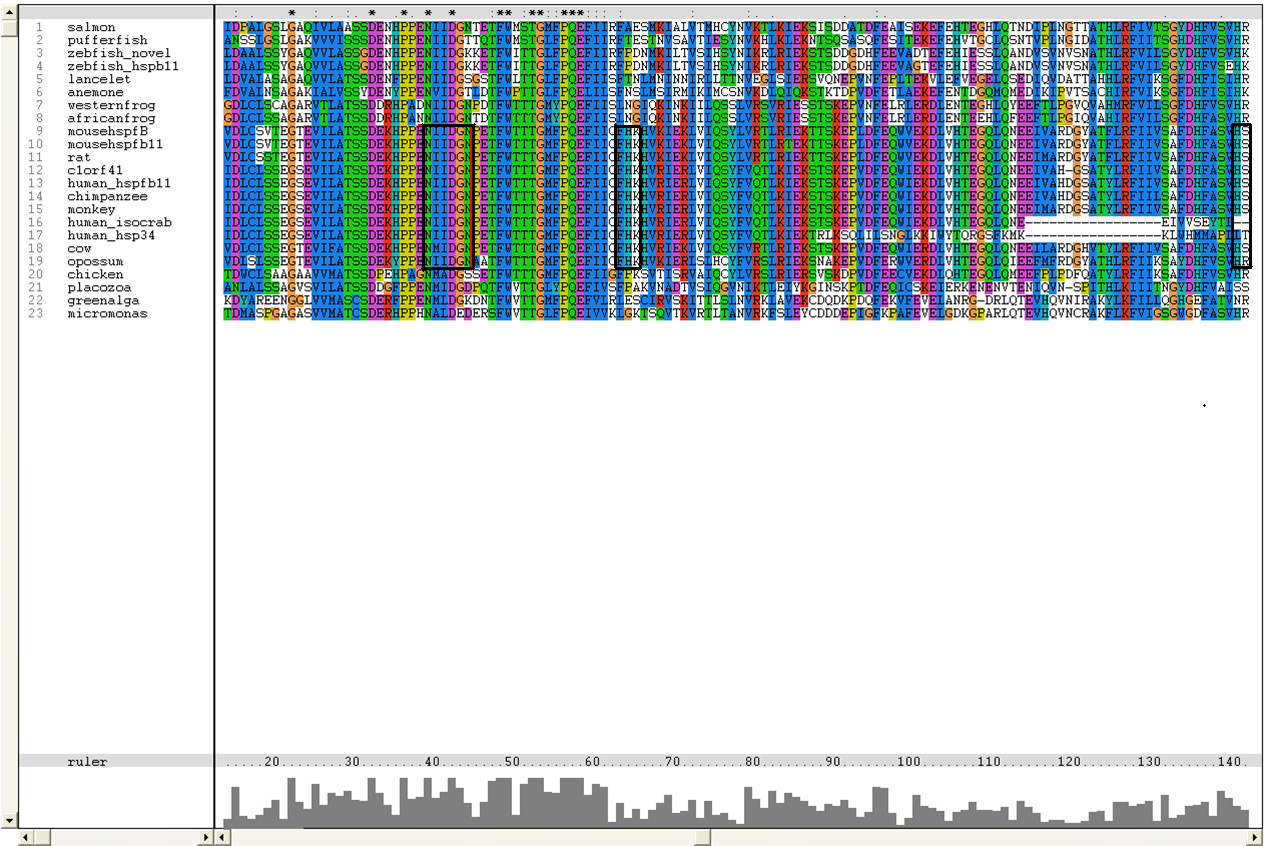
Multiple sequence alignment with all of the 27 sequences obtained from BLAST is shown below:
The sequence of platypus, urchin_156 and urchin_159 was removed due to poor alignment. Besides that, the sequence for g.alga_fap was removed as it was a subsequence of g.alga_itp25.
Phylogenetic Tree
After obtaining the multiple sequence alignment, phylogenetic tree was construct based on the result from multiple sequence alignment. Bootstrap values are shown on each branch.