Chromosome 1 open reading frame 41 Evolution: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:BLAST.png|centre|framed|'''Figure 1:''' BLAST result]] | |||
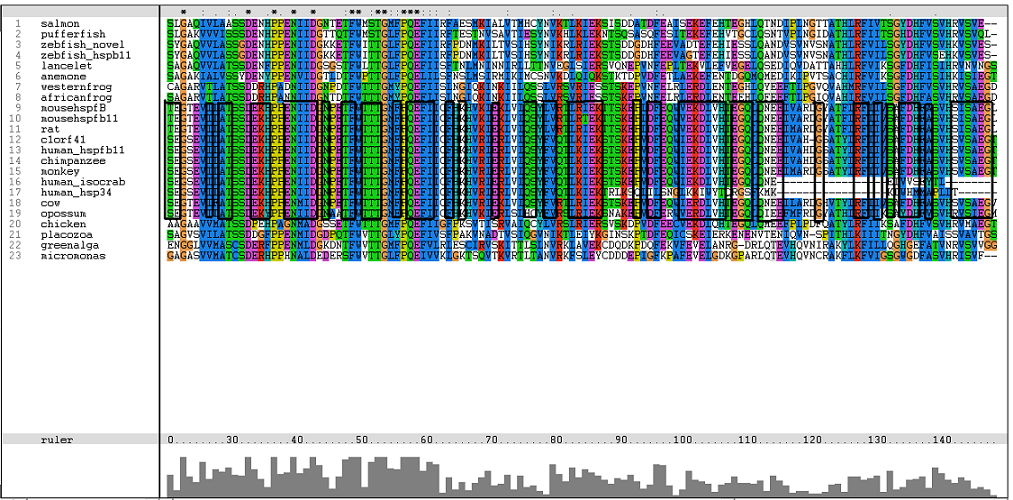
H-(2x)-W-(29x)-R-(2x)-S-(2x)-E residues critical for galactose binding site | |||
[[Image:Msa16.png|centre|framed|'''Figure 2:''' Conserved region of alpha B domain.]] | |||
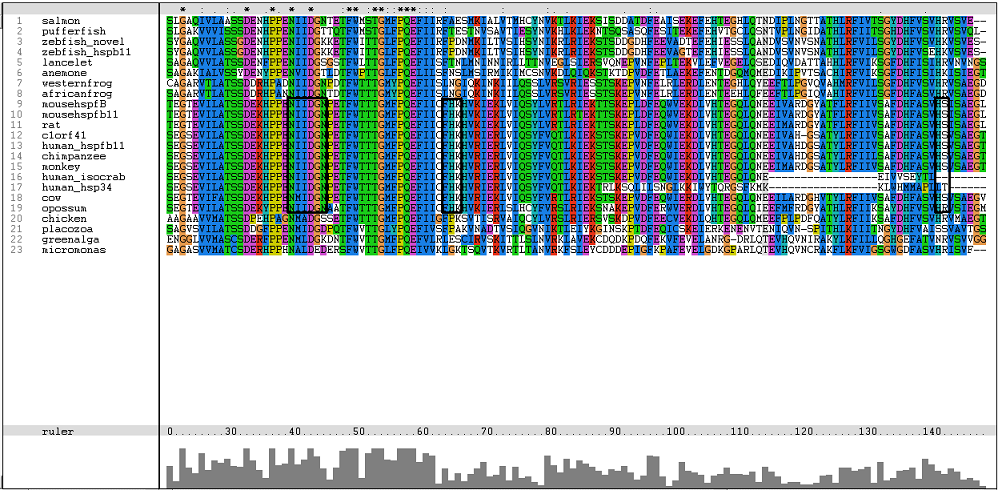
[[Image: | [[Image:Msa17.png|centre|framed|'''Figure 3:''' Residues involved in binding of Ca2+ and possible binding site.]] | ||
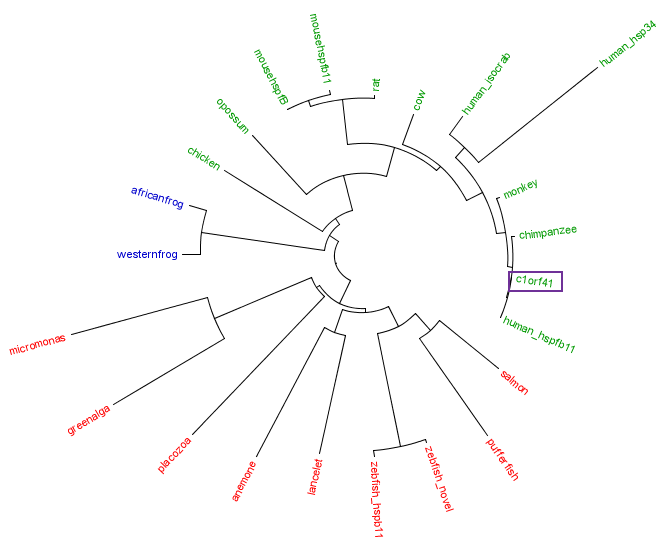
[[Image:Treecolour.png|centre|framed|'''Figure 4:''' Phylogenetic tree of c1orf41 with its homologues. Red is marine eukaryote, blue is amphibian and green is terrestrial eukaryotes.]] | |||
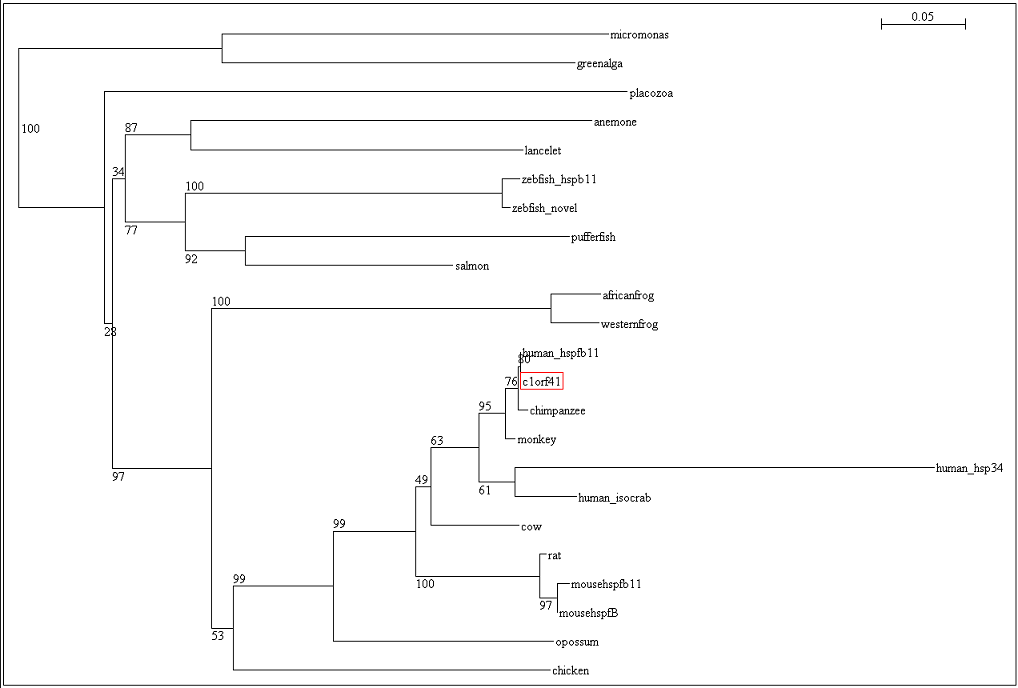
[[Image:Tree.png|centre|framed|'''Figure 5:''' Phylogenetic tree with bootstrap values]] | |||
[[Chromosome 1 open reading frame 41 Function|Next]] | |||
[[ | |||
Latest revision as of 06:26, 16 June 2009
H-(2x)-W-(29x)-R-(2x)-S-(2x)-E residues critical for galactose binding site