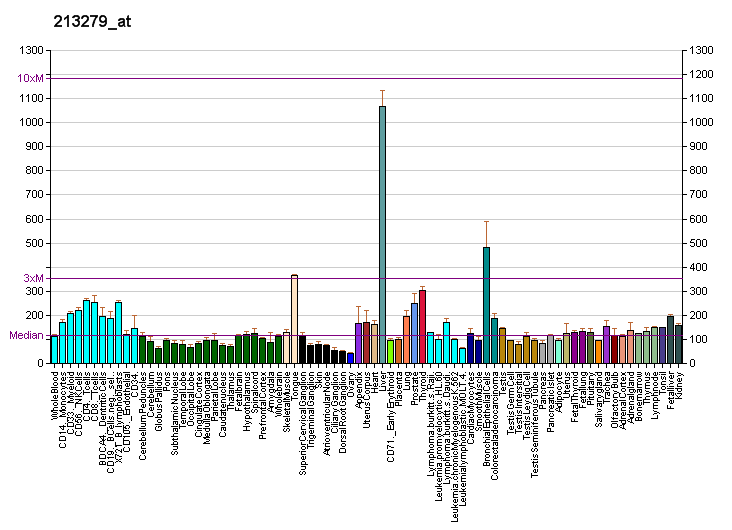
DHRS1 Results: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
[[Image:align 2uvd.png|framed|'''Figure 2'''<BR>Cartoon of DHRS1 (cyan/magenta) aligned with Oxoacyl-(Acyl-Carrier-protien)(green/red) the key catalytic Tyrosine residue is shown in as well. |none]]<BR> | [[Image:align 2uvd.png|framed|'''Figure 2'''<BR>Cartoon of DHRS1 (cyan/magenta) aligned with Oxoacyl-(Acyl-Carrier-protien)(green/red) the key catalytic Tyrosine residue is shown in as well. |none]]<BR> | ||
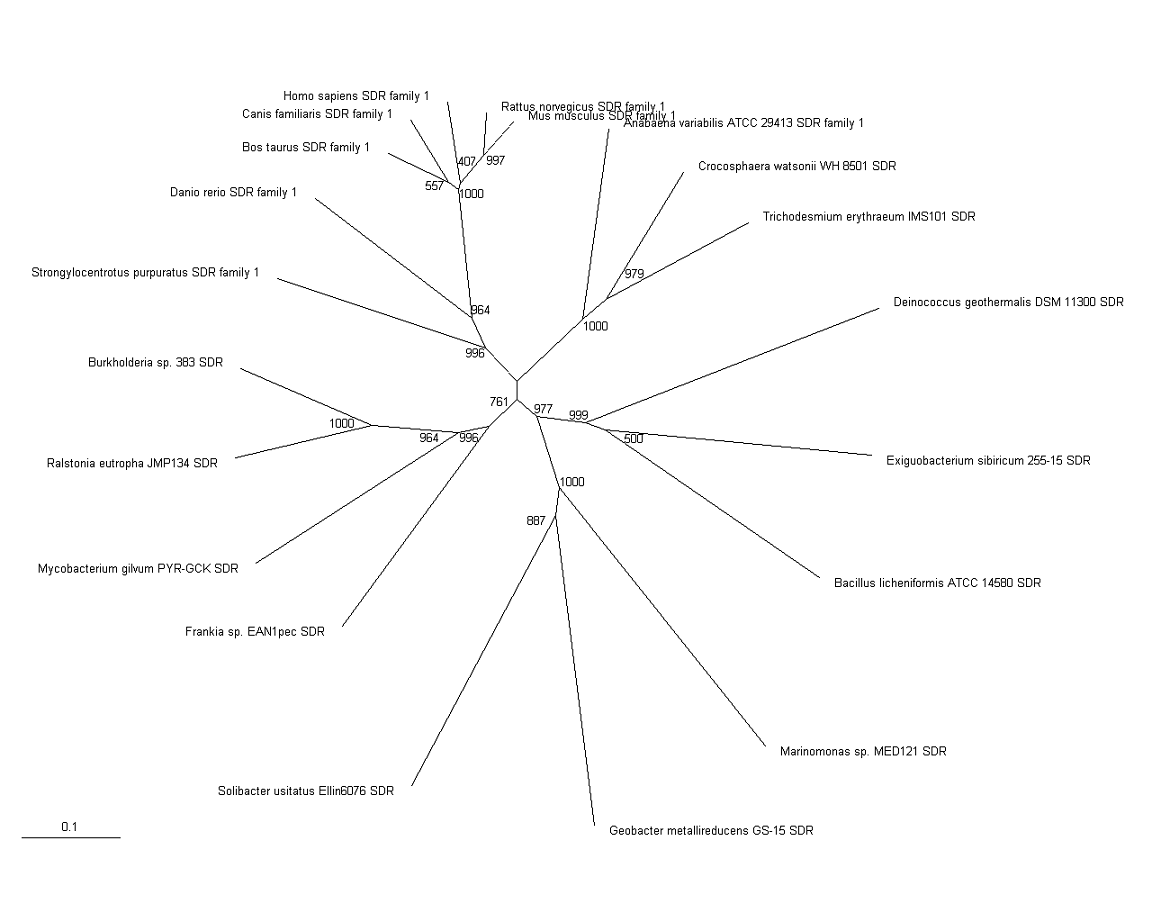
[[Image:StrappedTree.png| | [[Image:StrappedTree.png|framed|'''Figure 3'''<BR>Phylogenetic Tree of SDR Family with Bootstrapping values|1050px]]<BR> | ||
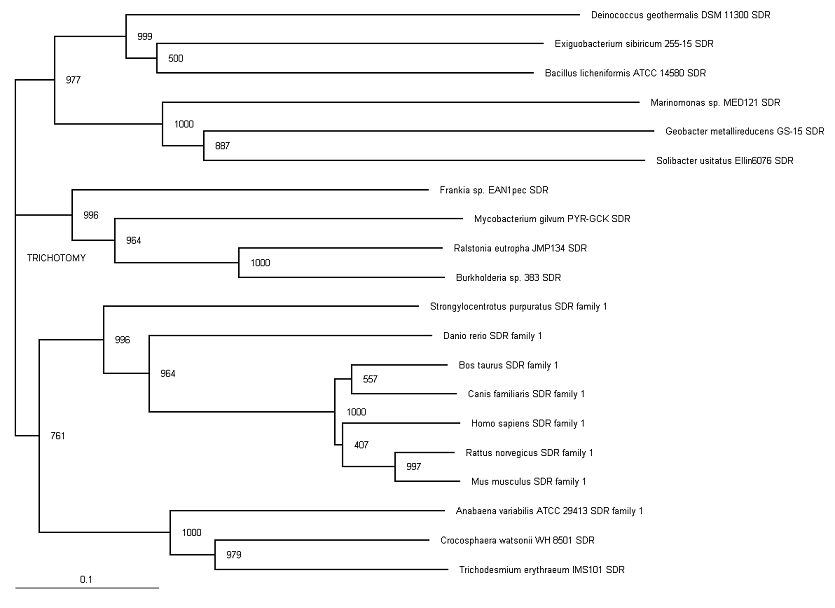
[[Image:Dehydrogenasereductase (SDR family) member 1 phylogram.png|framed|'''Figure 4'''<BR>Unrooted phylogram for Dehydrogenase/reductase (SDR family)|none]]<BR> | [[Image:Dehydrogenasereductase (SDR family) member 1 phylogram.png|framed|'''Figure 4'''<BR>Unrooted phylogram for Dehydrogenase/reductase (SDR family)|none]]<BR> | ||
Revision as of 08:51, 20 May 2008
Table 1
PDB/chain identifiers and structural alignment statistics for DALI search
No: Chain Z rmsd lali nres %id Description 1: 2qq5-A 48.1 0.0 238 238 100 MOL:DEHYDROGENASE/REDUCTASE SDR1; 2: 2uvd-A 29.6 2.1 220 246 27 MOL: 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE; 3: 1yde-F 29.3 2.1 216 256 29 MOL: RETINAL DEHYDROGENASE/REDUCTASE 3; 4: 1vl8-B 29.1 2.0 220 252 27 MOL: GLUCONATE 5-DEHYDROGENASE; 5: 2bgk-A 29.0 2.1 219 267 25 MOL: RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE; 6: 2q2q-D 28.9 2.0 217 255 26 MOL: BETA-D-HYDROXYBUTYRATEDEHYDROGENASE; 7: 1rwb-F 28.8 2.2 221 261 24 MOL: GLUCOSE 1-DEHYDROGENASE; 8: 1rwb-A 28.8 2.3 222 261 24 MOL: GLUCOSE 1-DEHYDROGENASE; 9: 1gee-A 28.8 2.3 222 261 24 MOL: GLUCOSE 1-DEHYDROGENASE; 10: 1gco-A 28.8 2.3 222 261 24 MOL: GLUCOSE DEHYDROGENASE; 11: 2zat-A 28.7 2.1 221 251 23 MOL: DEHYDROGENASE/REDUCTASE SDR4; 12: 1gee-B 28.7 2.3 221 261 24 MOL: GLUCOSE 1-DEHYDROGENASE; 13: 1gco-E 28.7 2.3 221 261 24 MOL: GLUCOSE DEHYDROGENASE;