Discussion of SNAPG: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
The SNAP-gamma structure obtained Danio rerio possess 4 chains of alpha helical hairpins fold without any beta strands observed. Each chain was interconnected with one another via hydrogen bond and non-bonded contacts (PDBSum). The helices towards N-terminal end were connected by extended loops and those towards C-terminal end tend to form a globular bundle presumably forming a cleft. Two different types of ligand interact with SNAPG differently. One of the ligand types, sulfanate ion (SO4), was found to from a distinct interaction with different residues in different chain (Figure 1 & 2). Meanwhile, selenomethione (MSE) ligands were integrated into protein structure as seem to appear as modified amino acid residues (Figure 1). | |||
Although the protein structure of SNAP-gamma was already known, the function is still yet under investigations. Protein super families identification reveals that SNAPG is made of 2 domains in which both of the domains had a soluble-NFS(N-ethylmaleimide-sensitive factor)-attachment-protein (SNAP) activity. The sequence conservation against the protein super families suggests that SNAP proteins may have a conserved 3D shape (need ref). Structural alignment against a representative structure in PDB databases using Dali web server shows several proteins with acknowledged protein function and domains shares structure similarity with SNAP-gamma protein (Table 1). For this study purpose, two proteins with highest statistical significance of the match (Z_value) were chosen. A vesicular transport protein sec17 found in yeast and type 4 fimbrial biogenesis protein pili in Pseudomonas aeruginosa had a Z-value of 23.3 and 12.9; and shares 23 and 14 respectively %identity with SNAP-gamma. Structure-based alignment of these structures indicates similar secondary structure and motifs (Figure 4&5). In regards of SNAP-gamma and sec17 surface properties proximity (Figure 6&7), it suggest that both of these proeteins may have similar protein-protein interaction. However, the amino acid conservation were not satisfying (Figure 5) suggesting the function can not be determined based on structure alignment alone. | |||
The SNAP-gamma hydrophobicity and electrostatic potential properties were distributed throughout majority of SNAP-gamma residues. These features may be important for examination of possible SNAP-gamma biological function. Large hydrophobic surface area, as shown by Figure 6.A, may contributes to ligands recognition and binding. Vast majority of SNAPG negative electrostatic potentials towards C-terminal end gave an insight of protein implausibility to form an interaction with DNA (Figure 9). This also supported by ProFunc results of which there was no HTH (helix-turn-helix) motif were found in the stucture. | |||
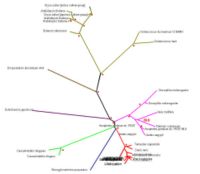
[[Image:Rad.JPG|thumb|200px|Figure 3. Evolutionary analysis of the tree with radial view. Red dotted signs means bootstrap value more than 75% or 75.00|right]] | [[Image:Rad.JPG|thumb|200px|Figure 3. Evolutionary analysis of the tree with radial view. Red dotted signs means bootstrap value more than 75% or 75.00|right]] | ||
Revision as of 23:28, 11 June 2007
The SNAP-gamma structure obtained Danio rerio possess 4 chains of alpha helical hairpins fold without any beta strands observed. Each chain was interconnected with one another via hydrogen bond and non-bonded contacts (PDBSum). The helices towards N-terminal end were connected by extended loops and those towards C-terminal end tend to form a globular bundle presumably forming a cleft. Two different types of ligand interact with SNAPG differently. One of the ligand types, sulfanate ion (SO4), was found to from a distinct interaction with different residues in different chain (Figure 1 & 2). Meanwhile, selenomethione (MSE) ligands were integrated into protein structure as seem to appear as modified amino acid residues (Figure 1).
Although the protein structure of SNAP-gamma was already known, the function is still yet under investigations. Protein super families identification reveals that SNAPG is made of 2 domains in which both of the domains had a soluble-NFS(N-ethylmaleimide-sensitive factor)-attachment-protein (SNAP) activity. The sequence conservation against the protein super families suggests that SNAP proteins may have a conserved 3D shape (need ref). Structural alignment against a representative structure in PDB databases using Dali web server shows several proteins with acknowledged protein function and domains shares structure similarity with SNAP-gamma protein (Table 1). For this study purpose, two proteins with highest statistical significance of the match (Z_value) were chosen. A vesicular transport protein sec17 found in yeast and type 4 fimbrial biogenesis protein pili in Pseudomonas aeruginosa had a Z-value of 23.3 and 12.9; and shares 23 and 14 respectively %identity with SNAP-gamma. Structure-based alignment of these structures indicates similar secondary structure and motifs (Figure 4&5). In regards of SNAP-gamma and sec17 surface properties proximity (Figure 6&7), it suggest that both of these proeteins may have similar protein-protein interaction. However, the amino acid conservation were not satisfying (Figure 5) suggesting the function can not be determined based on structure alignment alone.
The SNAP-gamma hydrophobicity and electrostatic potential properties were distributed throughout majority of SNAP-gamma residues. These features may be important for examination of possible SNAP-gamma biological function. Large hydrophobic surface area, as shown by Figure 6.A, may contributes to ligands recognition and binding. Vast majority of SNAPG negative electrostatic potentials towards C-terminal end gave an insight of protein implausibility to form an interaction with DNA (Figure 9). This also supported by ProFunc results of which there was no HTH (helix-turn-helix) motif were found in the stucture.
nanti aku mau jelasin ttg ligandnya
Tea: evolution linked with function.
SNAP-gamma protein evolutionary tree mostly contains more than 75% boostrapvalue. Thus, red dot signs was assign to represent more than 75% boostrap value. The value was indicated the degree of confidence about the branching order. As an example: in the diagram phylogenetic tree (see Figure 3), there is 39.0 boostrap value in branching Apis melifera and Tribolium castaneum. The boostrap value is below 75%, thus it could be concluded that the degree of confidence of branching tree in the correct order is far below 95%. On the other hand, boostrap value more than 75% or 75.00 means 95% confidence that the branching tree is in the correct order
As indicated in diagram in Figure 3, SNAP-gamma protein was evolved in eukaryotes. There is no prokaryotes organism detected in the phylogenetic tree.
Function (in various organism) = nunggu justine
Thus, it could be concluded that SNAP-gamma protein is an important protein in eukaryotes animal.