Evolution: Difference between revisions
JasonCheong (talk | contribs) |
JasonCheong (talk | contribs) |
||
| Line 16: | Line 16: | ||
|} | |} | ||
'''Figure 1.''' '''Amino Acid Sequence (260 aa)''' of N- | '''Figure 1.''' '''Amino Acid Sequence (260 aa)''' of N-acetylneuraminic Acid (2gfh) protein from ''Mus musculus'' (House Mouse) | ||
=='''Multiple Sequence Alignments (msa)'''== | =='''Multiple Sequence Alignments (msa)'''== | ||
Revision as of 17:12, 10 June 2007
Query Sequences (2gfh)
| mgsdkihhhh hhmglsrvra vffdldntli dtagasrrgm levikllqsk yhykeeaeii
cdkvqvklsk ecfhpystci tdvrtshwee aiqetkggad nrklaeecyf lwkstrlqhm iladdvkaml telrkevrll lltngdrqtq rekieacacq syfdaivigg eqkeekpaps ifyhccdllg vqpgdcvmvg dtletdiqgg lnaglkatvw inksgrvplt sspmphymvs svlelpallq sidckvsmsv |
Figure 1. Amino Acid Sequence (260 aa) of N-acetylneuraminic Acid (2gfh) protein from Mus musculus (House Mouse)
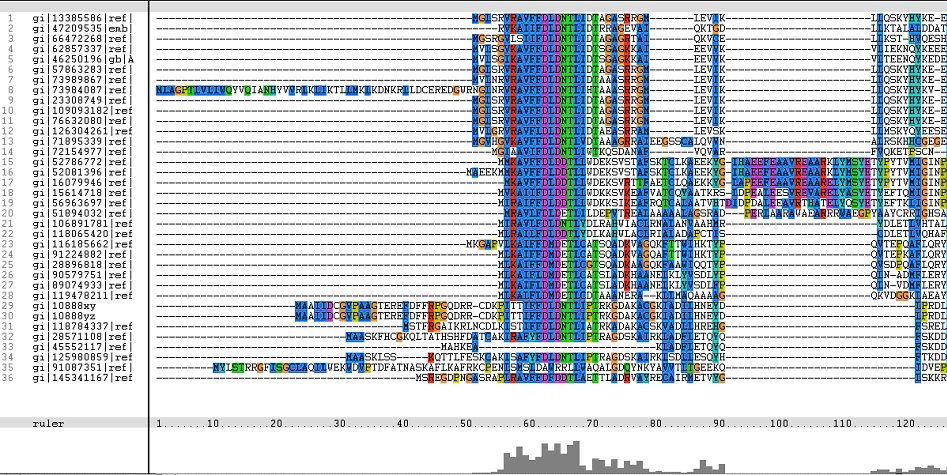
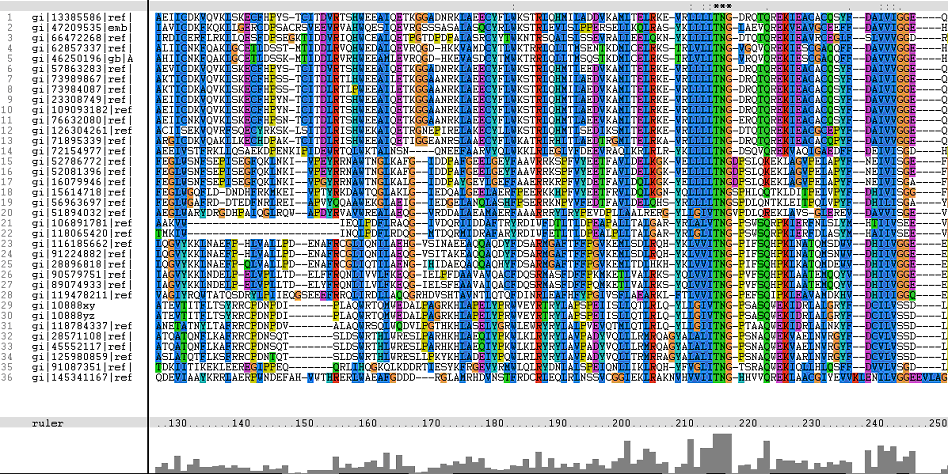
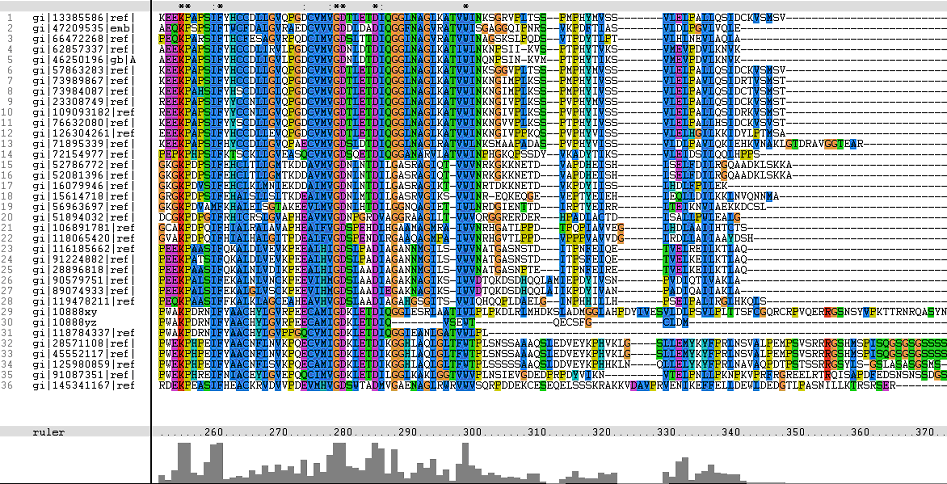
Multiple Sequence Alignments (msa)
Figure 1. Multiple Sequence Alignments of 2gfh amino acid sequences with that of other similar protein sequences in various organisms (A) Glycolysis pathway with substrates that are hydrolyze by HADs: glucose 6-phosphate, fructose 6-phosphate and dihydroxyacetone phosphate. (B) Pentose phostphate pathway with substrates that are hydrolyze by HADs: glucose-6-phosphate, fructose-6-phosphate, dihydroxyacetone phosphate, glyceraldehyde-3-phosphate, gluconate 6-phosphate and erythrose-4-phosphate.
| Catalytic activity | N-acylneuraminate 9-phosphate + H2O = N-acylneuraminate + phosphate |
| Cofactor | Magnesium (By similarity) |
| Enzyme regulation | Inhibited by vanadate and calcium (By similarity) |
| Pathway | Carbohydrate metabolism; aminosugar metabolism |
| Similarity | Belongs to the haloacid dehalogenase-like hydrolase superfamily. NANP family |