Evolution.: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| Line 22: | Line 22: | ||
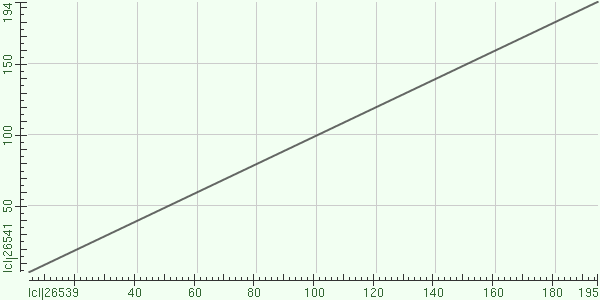
A dot matrix alignment between Drosophila (x-axis) and Human (y-axis) ssu72. The proteins are virtually identical. | A dot matrix alignment between Drosophila (x-axis) and Human (y-axis) ssu72. The proteins are virtually identical. | ||
[[Image: | [[Image:3fdfTree.jpg]] | ||
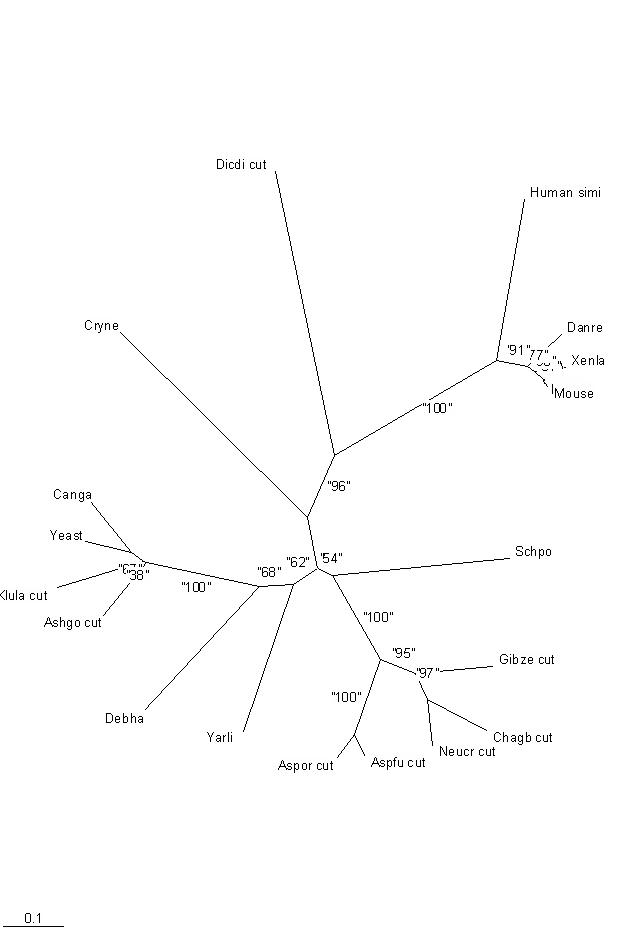
Tree, from SwissProt hits. | Tree, from SwissProt hits. | ||
[[Image:SSMPhosphatases.pdf]] | |||
Alignment of 3fdf (bottom sequence) to 7 phosphatases with similar secondary structure (top sequences). Conserved regions at 10 and 100, and occasional conserved residues. | |||
(Note phosphatases align much better with each other than with 3fdf) | |||
Revision as of 13:57, 11 June 2009
Multiple sequence alignments:
An alignment of a blast search of the NCBI's nonredundant database. Some of the sequences are protein or translated cDNA, but many come from genome annotations, with no functional information. The sources are both prokaryotes and eukaryotes.
This MSA shows that the sequence has been well conserved - some of the residues almost 100% conserved over a wide variety of organisms. However, if you use these to make other inferences, keep in mind that some of these sequences may not code for ssu72 (or anything else)...
An alignment from a blast search of SwissProt. The data is higher quality and less redundant, and there is functional information for each sequence. Of the 24 sequences, 3 are human ssu72 homologues, and the others are ssu72 from various eukaryotes. This MSA shows the conserved regions for ssu72. Since these are ssu72 sequences, I think the residues that show up as conserved here are probably important for structure/function.
Relation to other organisms
The taxonomy of the hits from SwissProt. Note these are all eukaryotes.
A dot matrix alignment between Drosophila (x-axis) and Human (y-axis) ssu72. The proteins are virtually identical.
Tree, from SwissProt hits.
Alignment of 3fdf (bottom sequence) to 7 phosphatases with similar secondary structure (top sequences). Conserved regions at 10 and 100, and occasional conserved residues. (Note phosphatases align much better with each other than with 3fdf)