Evolution.: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Image:SelNonredundantMSA.pdf]] | [[Image:SelNonredundantMSA.pdf]] | ||
| Line 7: | Line 5: | ||
This MSA shows that the sequence has been well conserved - some of the residues almost 100% conserved over a wide variety of organisms. However, if you use these to make other inferences, keep in mind that some of these sequences may not code for ssu72 (or anything else)... | This MSA shows that the sequence has been well conserved - some of the residues almost 100% conserved over a wide variety of organisms. However, if you use these to make other inferences, keep in mind that some of these sequences may not code for ssu72 (or anything else)... | ||
(H/V)C(X5)R(S/T) is very well conserved. | (H/V)C(X5)R(S/T) is very well conserved, as is the aspartate loop. | ||
[[Media:AnnotatedMSA2str.jpg]] | |||
[[Image:SwissProtMSA3.pdf]] | [[Image:SwissProtMSA3.pdf]] | ||
| Line 14: | Line 14: | ||
(Note: one of the sequences, 'CryneHomologue' is a homologue of the SwissProt 'Cryne' (Cryptococcus neoformans) sequence. The SwissProt sequence was a partial one, and ended up being too short.) | (Note: one of the sequences, 'CryneHomologue' is a homologue of the SwissProt 'Cryne' (Cryptococcus neoformans) sequence. The SwissProt sequence was a partial one, and ended up being too short.) | ||
(H/V)C(X5)R(S/T) is conserved in all of these sequences. | (H/V)C(X5)R(S/T) is conserved in all of these sequences. The aspartate loop is also well conserved. | ||
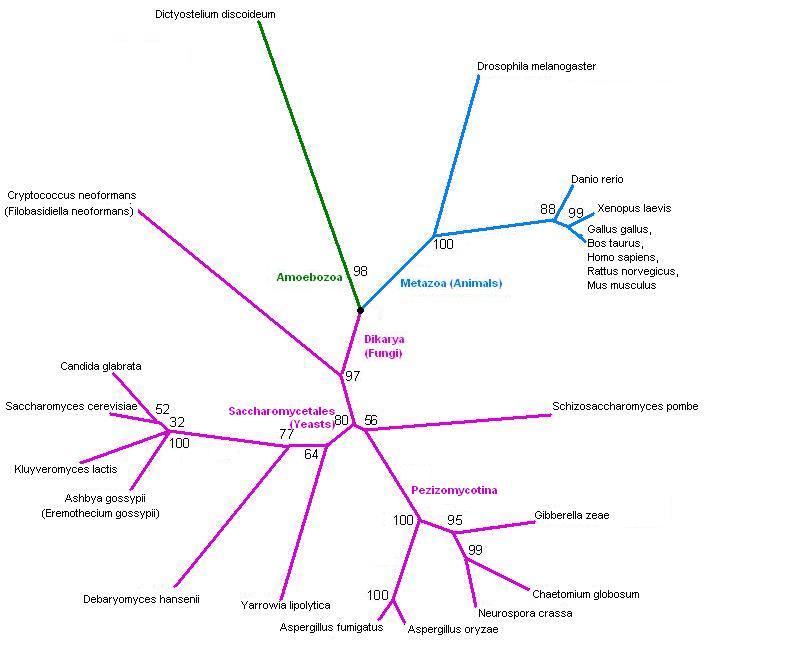
[[Image:3fdfTree2.jpg]] | [[Image:3fdfTree2.jpg]] | ||
Tree, from SwissProt hits. | Tree, from SwissProt hits. It splits into fungi and animals - and one slime mould. Drosophila is in with the animals, but fairly distant from human, as you'd expect. | ||
[[Image:SwissProtTax.txt]] | [[Image:SwissProtTax.txt]] | ||
| Line 28: | Line 26: | ||
[[Image:DrosophilaVsHuman.png]] | [[Image:DrosophilaVsHuman.png]] | ||
A dot matrix alignment between Drosophila (x-axis) and Human (y-axis) ssu72 | A dot matrix alignment between Drosophila (x-axis) and Human (y-axis) ssu72. | ||
[[Image:SSMPhosphatases.pdf]] | [[Image:SSMPhosphatases.pdf]] | ||
Latest revision as of 02:03, 16 June 2009
An alignment of a blast search of the NCBI's nonredundant database. Some of the sequences are protein or translated cDNA, but many come from genome annotations, with no functional information. The sources are both prokaryotes and eukaryotes.
This MSA shows that the sequence has been well conserved - some of the residues almost 100% conserved over a wide variety of organisms. However, if you use these to make other inferences, keep in mind that some of these sequences may not code for ssu72 (or anything else)...
(H/V)C(X5)R(S/T) is very well conserved, as is the aspartate loop.
An alignment from a blast search of SwissProt. The data is higher quality and less redundant. (Note: one of the sequences, 'CryneHomologue' is a homologue of the SwissProt 'Cryne' (Cryptococcus neoformans) sequence. The SwissProt sequence was a partial one, and ended up being too short.)
(H/V)C(X5)R(S/T) is conserved in all of these sequences. The aspartate loop is also well conserved.
Tree, from SwissProt hits. It splits into fungi and animals - and one slime mould. Drosophila is in with the animals, but fairly distant from human, as you'd expect.
The taxonomy of the hits from SwissProt. This confirms that the tree is showing the expected phylogeny.
A dot matrix alignment between Drosophila (x-axis) and Human (y-axis) ssu72.
Alignment of 3fdf (bottom sequence) to 7 phosphatases with similar secondary structure (top sequences). Conserved regions at 10 and 100, and occasional conserved residues. (Note phosphatases align much better with each other than with 3fdf)