Fascin Structure
Fascin structure and domains
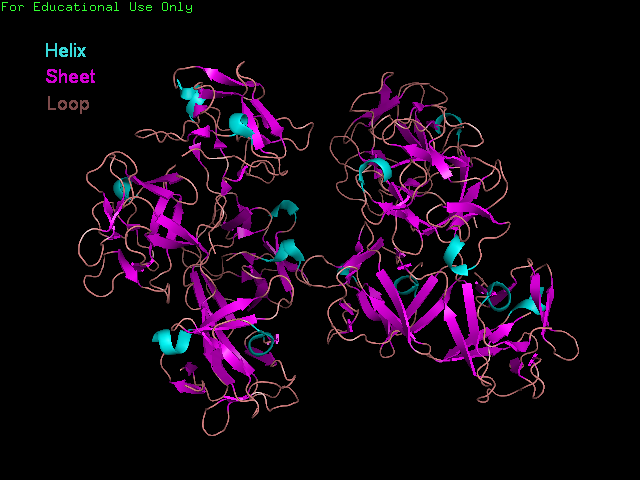
The Fascin protein is comprised of 8 repeats of the Fascin-like domain in total. It is made of two identical subgroups, each of which contains 4 Fascin-like domains linked together. The 2 subgroups are linked back to back slightly off-centre. The Fascin-like domain's tertiary structure adopts a beta-trefoil fold pattern with an internal threefold repeat. Secondary structure is predominantly Beta-sheet and random coil (loop) with small regions of alpha-helix. Structural similarity with several unrelated proteins is evident (eg. FGF).
'Front' view showing secondary structure of Fascin.
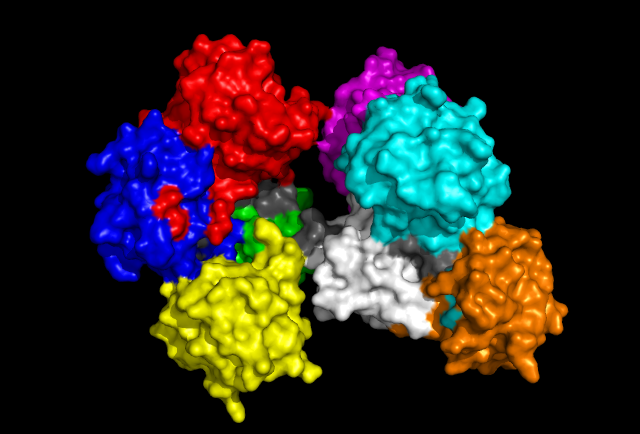
'Front' view of Fascin protein with each Fascin domain coloured. Red, Blue, Green and Yellow segments represent one subgroup while Aqua, Magenta, White and Orange represent the other. Grey colouration represents conserved residues found in sequence allignment.
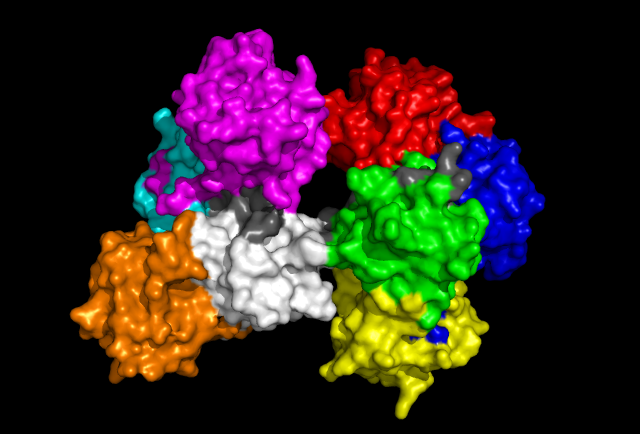
'Back' view of Fascin protein. Same colouration as above.
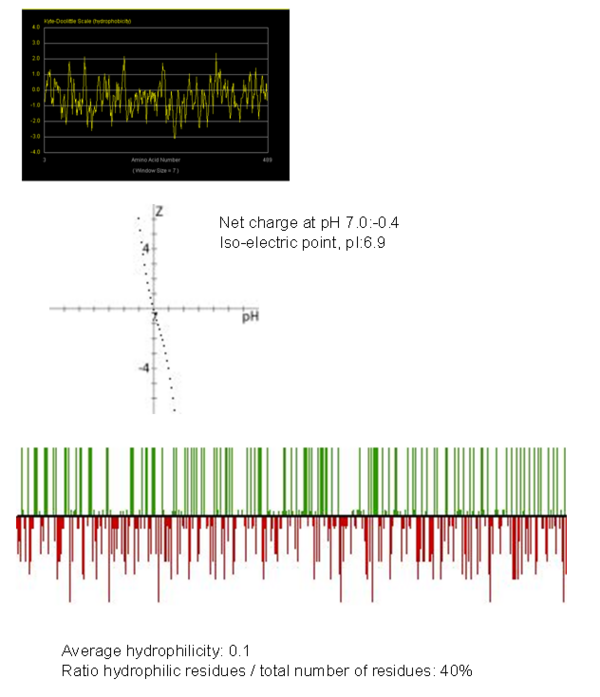
Hydrophobicity Plot
Plot showing the hydrophobicity plot of fascin-1 protein and second plot showing hydrophilicty plot which acts as a peptide property calculator. From these results we can overall state that this protein overall is hydrophobic.
Structual allignments
A Dali database search returned several hundred similar structures to Fascin 1. The first page of results can be seen below. Unfortunately none of the structurally similar molecules were actin binding so could not help with actin binding regions. The molecule 1ijt (Fibroblast Growth Factor - FGF) was alligned with Fascin to show structural similarity (even though they are functionally very different and only contain 9% sequence identity). Structurally FGF has much more Beta sheet secondarty structure, but as said before they are still quite similar.
Dali output showing first 33 structurally similar protein results.
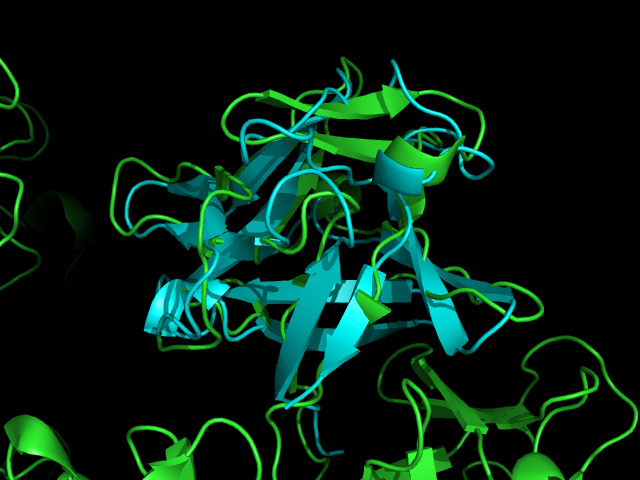
Pymol allignment of Fascin 1 (green) and FGF (aqua)
References
Joe Dundas, Zheng Ouyang, Jeffery Tseng, Andrew Binkowski, Yaron Turpaz, and Jie Liang 2006. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated resiudes.Nucleic Acid Research, 34:W116-W118.
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup, execution, and analysis of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res, 32, W665-W667 (2004).
Marchler-Bauer A et al. (2009), "CDD: specific functional annotation with the Conserved Domain Database.", Nucleic Acids Res. 37(D)205-10.