|
|
| Line 127: |
Line 127: |
| '''''Table 2.2 '''''Aligned sequence of 2ph1 protein | | '''''Table 2.2 '''''Aligned sequence of 2ph1 protein |
|
| |
|
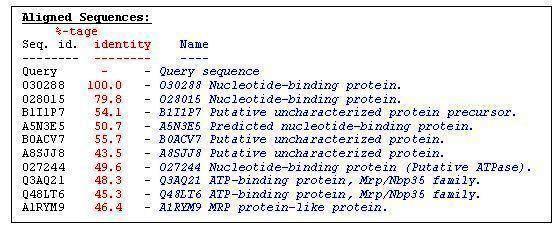
| '''Aligned Sequences:'''
| | [[Image:Ali.jpg]] |
| | |
| %-tage
| |
| | |
| Seq. id. identity Name
| |
| | |
| <nowiki>-------- </nowiki><nowiki>--------</nowiki> <nowiki>----</nowiki>
| |
| | |
| Query - - '''Query sequence '''
| |
| | |
| O30288 100.0 - '''O30288 Nucleotide-binding protein. '''
| |
| | |
| O28015 79.8 - '''O28015 Nucleotide-binding protein. '''
| |
| | |
| B1I1P7 54.1 - '''B1I1P7 Putative uncharacterized protein precursor. '''
| |
| | |
| A5N3E5 50.7 - '''A5N3E5 Predicted nucleotide-binding protein. '''
| |
| | |
| B0ACV7 55.7 - '''B0ACV7 Putative uncharacterized protein. '''
| |
| | |
| A8SJJ8 43.5 - '''A8SJJ8 Putative uncharacterized protein. '''
| |
| | |
| O27244 49.6 - '''O27244 Nucleotide-binding protein (Putative ATPase). '''
| |
| | |
| Q3AQ21 48.3 - '''Q3AQ21 ATP-binding protein, Mrp/Nbp35 family. '''
| |
| | |
| Q48LT6 45.3 - '''Q48LT6 ATP-binding protein, Mrp/Nbp35 family. '''
| |
| | |
| A1RYM9 46.4 - '''A1RYM9 MRP protein-like protein. '''
| |
| | |
|
| |
|
|
| |
|
| Line 166: |
Line 137: |
|
| |
|
| {| class="prettytable" | | {| class="prettytable" |
| | [[Image:]] | | | [[Image:3seq1.jpg]] |
|
| |
|
| |- | | |- |
| Line 172: |
Line 143: |
|
| |
|
| |- | | |- |
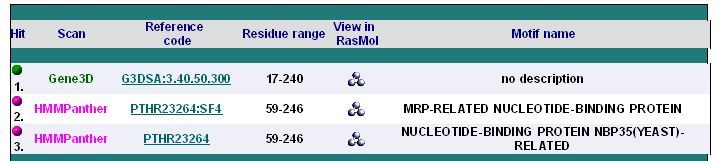
| | [[Image:|thumb|{| class="prettytable" | colspan="6" | <center></center> | |- | <center>'''Hit'''</center> | <center>'''Scan'''</center> | <center>'''Reference code'''</center> | <center>'''Residue range'''</center> | <center>'''View in RasMol'''</center> | <center>'''Motif name'''</center> | colspan="0" | | |- | colspan="6" | <center></center> | |- | <center>''' 1.'''</center> | <center>'''Gene3D'''</center> | <center>[http://gene3d.biochem.ucl.ac.uk/Gene3D/Results/quick_search?results=go&results=features&results=kegg&results=cog&results=interactions&search_term=G3DSA:3.40.50.300 G3DSA:3.40.50.300]</center> | <center>'''17-240'''</center> | <center></center> | <center>'''no description'''</center> | colspan="0" | | |- | <center>''' 2.'''</center> | <center>'''HMMPanther'''</center> | <center>[http://panther.appliedbiosystems.com/panther/family.do?clsAccession=PTHR23264:SF4 PTHR23264:SF4]</center> | <center>'''59-246'''</center> | <center></center> | <center>'''MRP-RELATED NUCLEOTIDE-BINDING PROTEIN'''</center> | colspan="0" | | |- | <center>''' 3.'''</center> | <center>'''HMMPanther'''</center> | <center>[http://panther.appliedbiosystems.com/panther/family.do?clsAccession=PTHR23264 PTHR23264]</center> | <center>'''59-246'''</center> | <center></center> | <center>'''NUCLEOTIDE-BINDING PROTEIN NBP35(YEAST)-RELATED'''</center> | colspan="0" | | |- | colspan="6" | <center></center> | |}]] | | | [[Image:|3seq2.jpg|{| class="prettytable" | colspan="6" | <center></center> | |- | <center>'''Hit'''</center> | <center>'''Scan'''</center> | <center>'''Reference code'''</center> | <center>'''Residue range'''</center> | <center>'''View in RasMol'''</center> | <center>'''Motif name'''</center> | colspan="0" | | |- | colspan="6" | <center></center> | |- | <center>''' 1.'''</center> | <center>'''Gene3D'''</center> | <center>[http://gene3d.biochem.ucl.ac.uk/Gene3D/Results/quick_search?results=go&results=features&results=kegg&results=cog&results=interactions&search_term=G3DSA:3.40.50.300 G3DSA:3.40.50.300]</center> | <center>'''17-240'''</center> | <center></center> | <center>'''no description'''</center> | colspan="0" | | |- | <center>''' 2.'''</center> | <center>'''HMMPanther'''</center> | <center>[http://panther.appliedbiosystems.com/panther/family.do?clsAccession=PTHR23264:SF4 PTHR23264:SF4]</center> | <center>'''59-246'''</center> | <center></center> | <center>'''MRP-RELATED NUCLEOTIDE-BINDING PROTEIN'''</center> | colspan="0" | | |- | <center>''' 3.'''</center> | <center>'''HMMPanther'''</center> | <center>[http://panther.appliedbiosystems.com/panther/family.do?clsAccession=PTHR23264 PTHR23264]</center> | <center>'''59-246'''</center> | <center></center> | <center>'''NUCLEOTIDE-BINDING PROTEIN NBP35(YEAST)-RELATED'''</center> | colspan="0" | | |- | colspan="6" | <center></center> | |}]] |
|
| |
|
| |} | | |} |
| Line 180: |
Line 151: |
|
| |
|
| {| class="prettytable" | | {| class="prettytable" |
| | [[Image:]] | | | [[Image:seq1.jpg]] |
|
| |
|
| |- | | |- |
| Line 186: |
Line 157: |
|
| |
|
| |- | | |- |
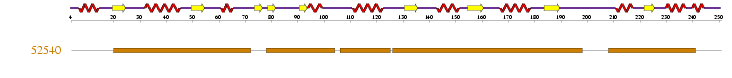
| | [[Image:|thumb|{| class="prettytable" | colspan="7" | |- | <div align="right">''' Hit '''</div> | <center>''' Reference code '''</center> | <center>''' Residue range '''</center> | <center>''' E-value '''</center> | <center>''' View in RasMol '''</center> | <center>''' PDB entry '''</center> | '''Name ''' |- | colspan="7" | |- | <div align="right">''' 1 '''</div> | <center>[http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?sunid=52540 52540] '''</center> | <center>'''20-7178-103106-124126-197208-240'''</center> | <center>'''4.02e-51 '''</center> | <center>''' '''</center> | <center>[http://www.ebi.ac.uk/pdbsum/1gm5 1gm5]A (d4) '''</center> | '''P-loop containing nucleoside triphosphate hydrolases ''' |- | colspan="7" | |}]] | | | [[Image:|seq2.jpg|{| class="prettytable" | colspan="7" | |- | <div align="right">''' Hit '''</div> | <center>''' Reference code '''</center> | <center>''' Residue range '''</center> | <center>''' E-value '''</center> | <center>''' View in RasMol '''</center> | <center>''' PDB entry '''</center> | '''Name ''' |- | colspan="7" | |- | <div align="right">''' 1 '''</div> | <center>[http://supfam.org/SUPERFAMILY/cgi-bin/scop.cgi?sunid=52540 52540] '''</center> | <center>'''20-7178-103106-124126-197208-240'''</center> | <center>'''4.02e-51 '''</center> | <center>''' '''</center> | <center>[http://www.ebi.ac.uk/pdbsum/1gm5 1gm5]A (d4) '''</center> | '''P-loop containing nucleoside triphosphate hydrolases ''' |- | colspan="7" | |}]] |
|
| |
|
| |} | | |} |
| Line 192: |
Line 163: |
|
| |
|
|
| |
|
| <center>[[Image:]]</center> | | <center>[[Image:inter1.jpg]]</center> |
| | |
| | |
| | |
| {| class="prettytable"
| |
| | colspan="3" |
| |
| {| class="prettytable"
| |
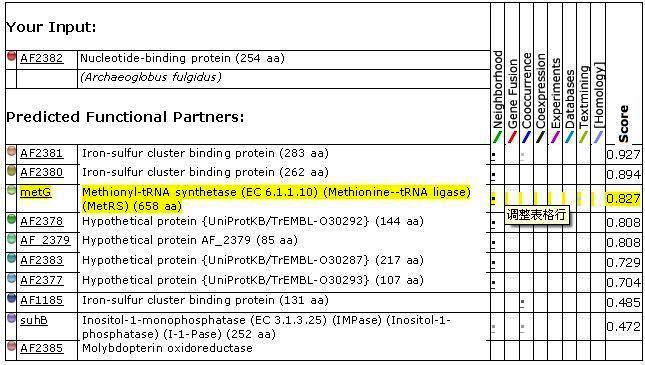
| | colspan="3" | '''Your Input:'''
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
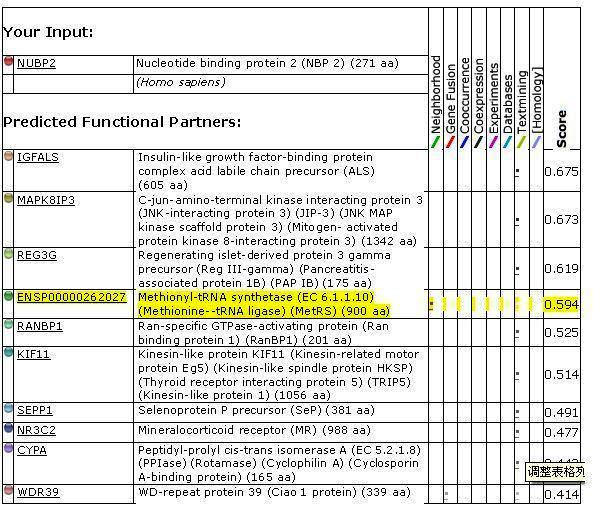
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900921&targetmode=proteins AF2382]
| |
| | Nucleotide-binding protein (254 aa)
| |
| | |
| |-
| |
| |
| |
| |
| |
| | ''(Archaeoglobus fulgidus)''
| |
| | |
| |-
| |
| | colspan="3" | '''Predicted Functional Partners:'''
| |
| | |
| |}
| |
| | |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900920&targetmode=proteins AF2381]
| |
| |
| |
| {| class="prettytable"
| |
| | Iron-sulfur cluster binding protein (283 aa)
| |
| | |
| |}
| |
| | |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900920 •]
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_ajax_phylo_evidence.pl?taskId=xYR99TnzadP2&node2=900920 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.927
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900919&targetmode=proteins AF2380]
| |
| |
| |
| {| class="prettytable"
| |
| | Iron-sulfur cluster binding protein (262 aa)
| |
| | |
| |}
| |
| | |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900919 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.894
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900002&targetmode=proteins metG]
| |
| |
| |
| {| class="prettytable"
| |
| | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS) (658 aa)
| |
| | |
| |}
| |
| | |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900002 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=xYR99TnzadP2&node2=900002 •]
| |
| |
| |
| | 0.827
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900917&targetmode=proteins AF2378]
| |
| |
| |
| {| class="prettytable"
| |
| | Hypothetical protein {UniProtKB/TrEMBL-O30292} (144 aa)
| |
| | |
| |}
| |
| | |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900917 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.808
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900918&targetmode=proteins AF_2379]
| |
| |
| |
| {| class="prettytable"
| |
| | Hypothetical protein AF_2379 (85 aa)
| |
| | |
| |}
| |
| | |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900918 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.808
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900922&targetmode=proteins AF2383]
| |
| |
| |
| {| class="prettytable"
| |
| | Hypothetical protein {UniProtKB/TrEMBL-O30287} (217 aa)
| |
|
| |
|
| |}
| |
|
| |
|
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900922 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.729
| |
|
| |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900916&targetmode=proteins AF2377]
| |
| |
| |
| {| class="prettytable"
| |
| | Hypothetical protein {UniProtKB/TrEMBL-O30293} (107 aa)
| |
|
| |
| |}
| |
|
| |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900916 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.704
| |
|
| |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=899737&targetmode=proteins AF1185]
| |
| |
| |
| {| class="prettytable"
| |
| | Iron-sulfur cluster binding protein (131 aa)
| |
|
| |
| |}
| |
|
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_ajax_phylo_evidence.pl?taskId=xYR99TnzadP2&node2=899737 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.485
| |
|
| |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900911&targetmode=proteins suhB]
| |
| |
| |
| {| class="prettytable"
| |
| | Inositol-1-monophosphatase (EC 3.1.3.25) (IMPase) (Inositol-1- phosphatase) (I-1-Pase) (252 aa)
| |
|
| |
| |}
| |
|
| |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=xYR99TnzadP2&node2=900911 •]
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_ajax_phylo_evidence.pl?taskId=xYR99TnzadP2&node2=900911 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | 0.472
| |
|
| |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=xYR99TnzadP2&node=900924&targetmode=proteins AF2385]
| |
| |
| |
| {| class="prettytable"
| |
| | Molybdopterin oxidoreductase
| |
|
| |
|
| |}
| |
|
| |
|
| |
| | <center>[[Image:input1.jpg]]</center> |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
|
| |
|
| |}
| |
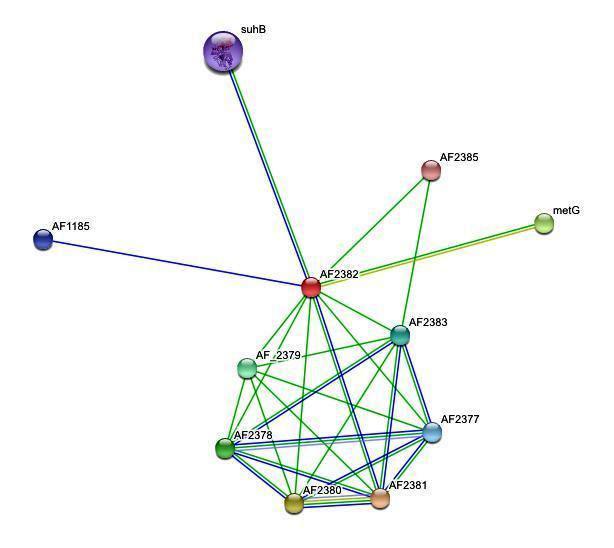
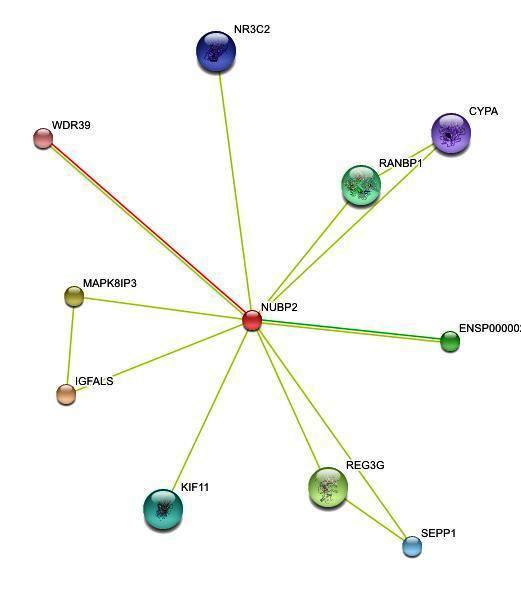
| '''''Figure 2. '''''Protein-protein interactions search using ''NUBP2 ''sequence | | '''''Figure 2. '''''Protein-protein interactions search using ''NUBP2 ''sequence |
|
| |
|
|
| |
|
| <center>[[Image:]]</center> | | <center>[[Image:inter2.jpg]]</center> |
| | |
| | |
| | |
| {| class="prettytable"
| |
| | colspan="3" |
| |
| {| class="prettytable"
| |
| | colspan="3" | '''Your Input:'''
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=310453&targetmode=proteins NUBP2]
| |
| | Nucleotide binding protein 2 (NBP 2) (271 aa)
| |
| | |
| |-
| |
| |
| |
| |
| |
| | ''(Homo sapiens)''
| |
| | |
| |-
| |
| | colspan="3" | '''Predicted Functional Partners:'''
| |
| | |
| |}
| |
| | |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | <center>[[Image:]]</center>
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=306506&targetmode=proteins IGFALS]
| |
| |
| |
| {| class="prettytable"
| |
| | Insulin-like growth factor-binding protein complex acid labile chain precursor (ALS) (605 aa)
| |
| | |
| |}
| |
| | |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=306506 •]
| |
| |
| |
| | 0.675
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=308840&targetmode=proteins MAPK8IP3]
| |
| |
| |
| {| class="prettytable"
| |
| | C-jun-amino-terminal kinase interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (1342 aa)
| |
| | |
| |}
| |
| | |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=308840 •]
| |
| |
| |
| | 0.673
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=312214&targetmode=proteins REG3G]
| |
| |
| |
| {| class="prettytable"
| |
| | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 1B) (PAP IB) (175 aa)
| |
| | |
| |}
| |
| | |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=312214 •]
| |
| |
| |
| | 0.619
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=310372&targetmode=proteins ENSP00000262027]
| |
| |
| |
| {| class="prettytable"
| |
| | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS) (900 aa)
| |
| | |
| |}
| |
| | |
| | [http://string.embl.de/newstring_cgi/show_neighborhood.pl?taskId=hKMxlIdcOMTb&node2=310372 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=310372 •]
| |
| |
| |
| | 0.594
| |
| | |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=320243&targetmode=proteins RANBP1]
| |
| |
| |
| {| class="prettytable"
| |
| | Ran-specific GTPase-activating protein (Ran binding protein 1) (RanBP1) (201 aa)
| |
| | |
| |}
| |
| | |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=320243 •]
| |
| |
| |
| | 0.525
| |
|
| |
|
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=310063&targetmode=proteins KIF11]
| |
| |
| |
| {| class="prettytable"
| |
| | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor interacting protein 5) (TRIP5) (Kinesin-like protein 1) (1056 aa)
| |
|
| |
|
| |}
| |
|
| |
|
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=310063 •]
| |
| |
| |
| | 0.514
| |
|
| |
|
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=327885&targetmode=proteins SEPP1]
| |
| |
| |
| {| class="prettytable"
| |
| | Selenoprotein P precursor (SeP) (381 aa)
| |
|
| |
| |}
| |
|
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=327885 •]
| |
| |
| |
| | 0.491
| |
|
| |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=325216&targetmode=proteins NR3C2]
| |
| |
| |
| {| class="prettytable"
| |
| | Mineralocorticoid receptor (MR) (988 aa)
| |
|
| |
| |}
| |
|
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=325216 •]
| |
| |
| |
| | 0.477
| |
|
| |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=325519&targetmode=proteins CYPA]
| |
| |
| |
| {| class="prettytable"
| |
| | Peptidyl-prolyl cis-trans isomerase A (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin A) (Cyclosporin A-binding protein) (165 aa)
| |
|
| |
| |}
| |
|
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=325519 •]
| |
| |
| |
| | 0.443
| |
|
| |
| |-
| |
| | <center>[[Image:]]</center>
| |
| | [http://string.embl.de/newstring_cgi/display_single_node.pl?taskId=hKMxlIdcOMTb&node=312224&targetmode=proteins WDR39]
| |
| |
| |
| {| class="prettytable"
| |
| | WD-repeat protein 39 (Ciao 1 protein) (339 aa)
| |
|
| |
| |}
| |
|
| |
|
| | | | -| <center>[[Image:input2.jpg]]</center> |
| | [http://string.embl.de/newstring_cgi/show_fusion_evidence.pl?taskId=hKMxlIdcOMTb&node2=312224 •]
| |
| |
| |
| |
| |
| |
| |
| |
| |
| | [http://string.embl.de/newstring_cgi/show_textmining_evidence.pl?taskId=hKMxlIdcOMTb&node2=312224 •]
| |
| |
| |
| | 0.414
| |
|
| |
|
| |} | | |} |
| Line 648: |
Line 185: |
|
| |
|
|
| |
|
| [[Image:]] | | [[Image:exp.jpg]] |