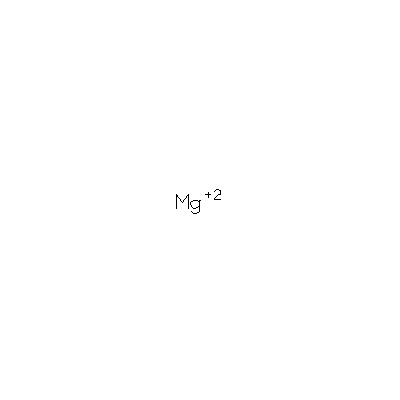Function for haloacid dehalogenase-like hydrolase domain containing 2: Difference between revisions
No edit summary |
No edit summary |
||
| Line 44: | Line 44: | ||
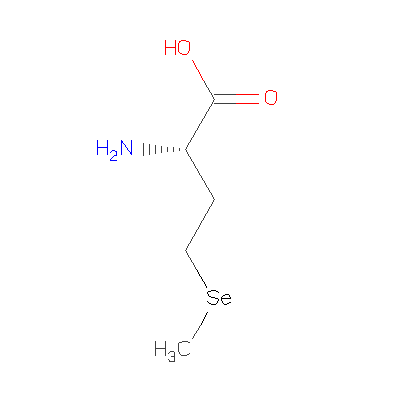
[http://www.rcsb.org/pdb/ligand/ligandsummary.do?handler=biologyChemistry&hetId=MSE / MSE] | [http://www.rcsb.org/pdb/ligand/ligandsummary.do?handler=biologyChemistry&hetId=MSE / MSE] | ||
[[Image:MSE 400.gif]] | [[Image:MSE 400.gif]] | ||
And at Mouse Genome Informatics (MGI) [http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=markerGO&key=60967 /Gene Ontology Classifications] | |||
We found 5 Category of Function or Process with evidence. | |||
<pre> | |||
Category Classification Term Evidence | |||
Molecular Function catalytic activity IEA | |||
Molecular Function hydrolase activity IEA | |||
Molecular Function hydrolase activity IEA | |||
Molecular Function magnesium ion binding IEA | |||
Biological Process metabolic process IEA | |||
Gene Ontology Evidence Code Abbreviations: | |||
IEA = Inferred from electronic annotation | |||
</pre> | |||
Revision as of 02:40, 5 June 2007
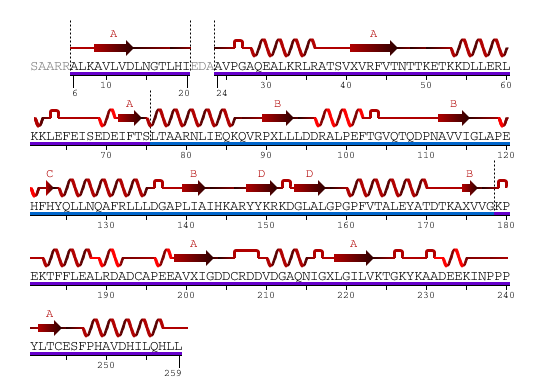
Haloacid dehalogenase-like hydrolase domain containing 2 (2HO4:A,B)
Where's does this name come from? We look at it's name.
Hydolase indicates it's main molecular function, hydrolase activity.
And, Domain containing 2 illustrate it has 2 domains / Sequence and Secondary Structure
From the above diagram, we see 2 domain color in blue and purple showed separately.
Type: polypeptide(L)
Length: 259 residues
Secondary Structure: 44% helical (14 helices; 123 residues) 17% beta sheet (12 strands; 48 residues)
2HO4Aa: pdp domain 2HO4Aa (in purple)
2HO4Ab: pdp domain 2HO4Ab (in blue)
But this does not conclude all it's function
From the Protein Data Bank (PDB) /PBD Biology and Chemistry Report
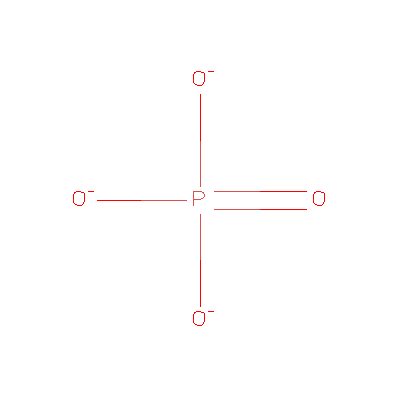
We found it has 3 Ligands and Prosthetic Groups
ID Name Chemical Formula Weight Ligand PO4 Phosphate Ion O4 P 3- 94.971 MG Magnesium Ion Mg 2+ 24.305 MSE Selenomethionine C5 H11 N O2 Se 196.107
And at Mouse Genome Informatics (MGI) /Gene Ontology Classifications
We found 5 Category of Function or Process with evidence.
Category Classification Term Evidence Molecular Function catalytic activity IEA Molecular Function hydrolase activity IEA Molecular Function hydrolase activity IEA Molecular Function magnesium ion binding IEA Biological Process metabolic process IEA Gene Ontology Evidence Code Abbreviations: IEA = Inferred from electronic annotation
CDS 1..259
/gene="HDHD2"
/coded_by="NM_032124.4:134..913"
/GO_function="hydrolase activity"
/GO_process="metabolism"
/db_xref="CCDS:CCDS32829.1"
/db_xref="GeneID:84064"
/db_xref="HGNC:25364"
/db_xref="HPRD:13640"
GO_function:/ hydrolase activity
GO_process:/ metabolic process
Gene Detail:/ Hdhd2
FASTA of HDHD2 of haloacid dehalogenase-like hydrolase domain containing 2 Homo sapiens
COExpresson of Predict Functional Protein:/ HDHD2 haloacid dehalogenase-like hydrolase domain containing 2
ProFunc Result:/ 2ho4
HDHD2 (259 aa) (Homo sapiens) is highly conservatity to Pan troglodytes, Macaca mulatta, Rattus norvegicus, Mus musculus species.
Predicted Functional Partners: KIAA0423 annotation not available (1723 aa)
/ Gene Ontology Classifications