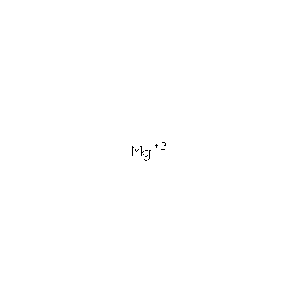Function for haloacid dehalogenase-like hydrolase domain containing 2
Haloacid dehalogenase-like hydrolase domain containing 2 (2HO4:A,B)
Where's does this name come from?
Hydolase indicates it's main molecular function, hydrolase activity. /Link
Definition of hydrolase activity:
Catalysis of the hydrolysis of various bonds
e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc.
Hydrolase is the systematic name for any enzyme of EC class 3.
The name Domain containing 2 illustrate it has 2 domains
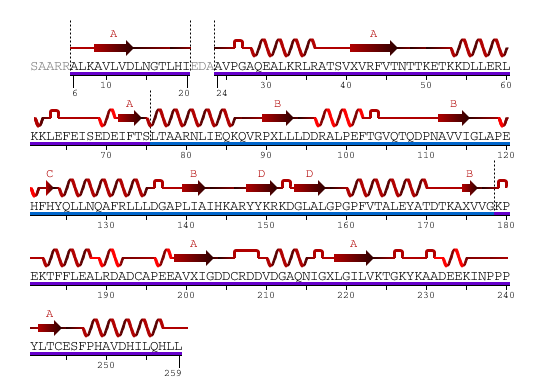
Sequence and Secondary Structure /Link
From the above diagram, we see 2 domains color in blue and purple showed separately.
Type: polypeptide(L)
Length: 259 residues
Secondary Structure: 44% helical (14 helices; 123 residues) 17% beta sheet (12 strands; 48 residues)
2HO4Aa: pdp domain 2HO4Aa (in purple)
2HO4Ab: pdp domain 2HO4Ab (in blue)
But this does not conclude all it's function
Protein Data Bank (PDB)
Ligands and Prosthetic Groups
| ID | Name Chemical | Formula | Weight Ligand | Link |
|---|---|---|---|---|
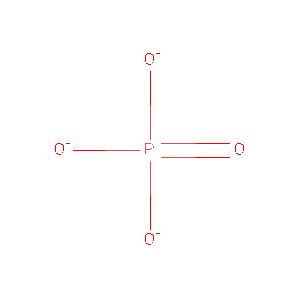
| PO4 | Phosphate Ion | O4 P 3- | 94.971 | /View |
| MG | Magnesium Ion | Mg 2+ | 24.305 | /View |
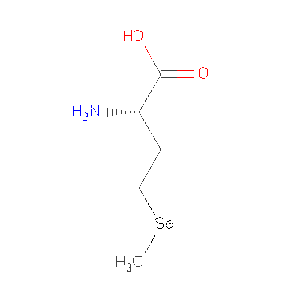
| MSE | Selenomethionine | C5 H11 N O2 Se | 196.107 | /View |
PBD Biology and Chemistry Report /Link
Mouse Genome Informatics(MGI)
Gene Ontology Classifications /Link
We found 5 Category of Function or Process with evidence.
GO Annotations in Tabular Form
| Category | Classification | Term Evidence | Inferred From | Ref(s) |
|---|---|---|---|---|
| Molecular | Function catalytic activity | IEA | InterPro:IPR005834 | J:72247 |
| Molecular Function | hydrolase activity | IEA | InterPro:IPR006357 | J:72247 |
| Molecular Function | hydrolase activity | IEA | SP_KW:KW-0378 | J:60000 |
| Molecular Function | magnesium ion binding | IEA | SP_KW:KW-0460 | J:60000 |
| Biological Process | metabolic process | IEA | InterPro:IPR005834,InterPro:IPR006357 | J:72247 |
Gene Ontology Evidence Code Abbreviations: IC Inferred by curator IDA Inferred from direct assay IEA Inferred from electronic annotation IGI Inferred from genetic interaction IMP Inferred from mutant phenotype IPI Inferred from physical interaction ISS Inferred from sequence or structural similarity NAS Non-traceable author statement ND No biological data available RCA Reviewed computational analysis TAS Traceable author statement
So where's the evidence from?
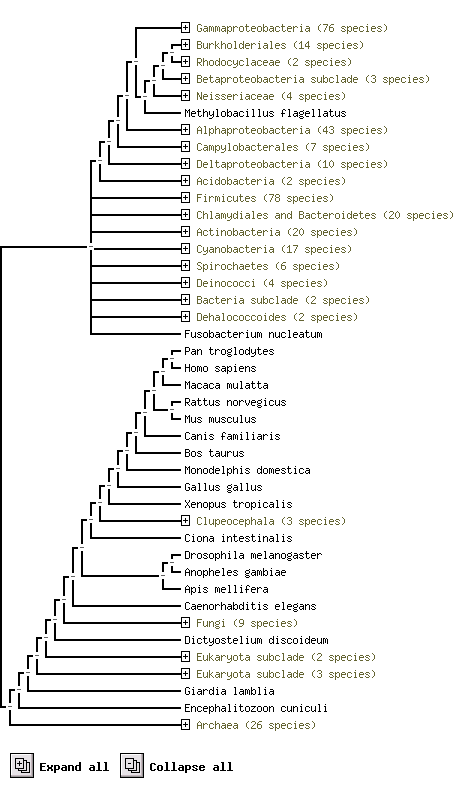
The evidence was done by Mammalian Orthology Load, which information was derived from Homologene National Center for Biotechnology Information. This polymer was reviewed in 2004 and record with J Number: J:90500.
The Mammalian Orthology Query Results shows the Orthology Evidence were used both Amino acid sequence comparison and NT Nucleotide sequence comparison. With species of human, mouse(laboratory), rat, chimpanzee and dog(domestic) all contain this polymer.
What about it's functional partners
From /STRING uses orthology information from the excellent COG database, we predicted it's functional partners.
From the result of occurrence we discover KIAA0423 annotation, which is still not available (1723 aa) is it best suit predicted Functional Partners. / more information
The above result shows using protein sequence of ploymer to compare in difference species and their sequence conservation.
e.g. Pan troglodytes and Homo sapiens are two species with this polymer similiar
How does hydrolase activity?
The HAD (haloacid dehalogenase) superfamily of hydrolases contains enzymes such as L-2-haloacid dehalogenase, epoxide hydrolase and a variety of phosphatases
HDHD2 (259 aa) (Homo sapiens) is highly conservatity to Pan troglodytes, Macaca mulatta, Rattus norvegicus, Mus musculus species.
Predicted Functional Partners: KIAA0423 annotation not available (1723 aa)
/ Gene Ontology Classifications
/ Mammalian Orthology Proteins
http://symatlas.gnf.org/SymAtlas/symquery?q=212508