Functional analysis of 2ece: Difference between revisions
From MDWiki
Jump to navigationJump to search
No edit summary |
|||
| Line 4: | Line 4: | ||
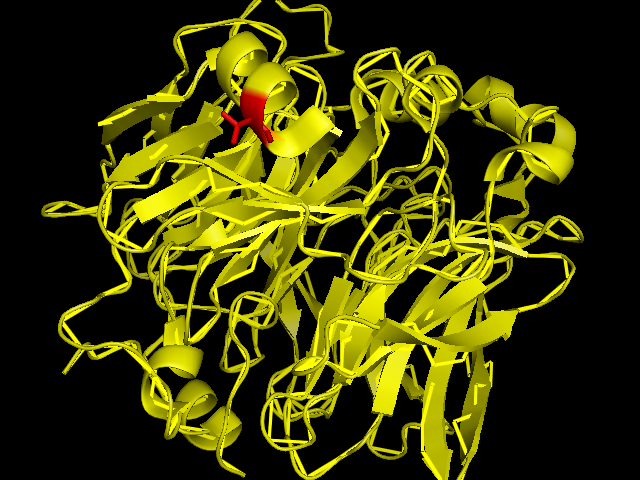
[[Image:SBP with Sele atom.png|frame|border|left|The red coloured region is the region carrying Se, and it is evident that the Se is incorporated in an amino acid of the helix chain.(Pymol Image) | [[Image:SBP with Sele atom.png|frame|border|left|The red coloured region is the region carrying Se, and it is evident that the Se is incorporated in an amino acid of the helix chain.(Pymol Image) | ||
]] | ]] | ||
| Line 117: | Line 107: | ||
The above table shows some condensed Dali results. The proteins that might have direct carcinogenic effects via the immune system are highlighted. Most of the proteins that have structural similarity to SBP are highly involved in redox reactions,like the reductases. | |||
Revision as of 01:04, 10 June 2008
FUNCTIONAL analysis of SELENBP 1 (pdb 2ece)
DaliLite: Structural Neighbours
Query: mol1A MOLECULE: 462AA LONG HYPOTHETICAL SELENIUM-BINDING PROTEIN;
Matches are sorted by Z-score. Similarities with a Z-score lower than 2 are spurious.
Summary
No: |Chain| Z| rmsd| lali| nres| %id| Description
1: |2ece-A |80.1 |0.0 |455 |455 |100 |MOLECULE: 462AA LONG HYPOTHETICAL SELENIUM-BINDING PROTEIN; 2: |1ri6-A |32.6 |2.6 |314 |333 | 12 |MOLECULE: PUTATIVE ISOMERASE YBHE; 3: |1jof-A |31.3 |2.9 |329 |365 |10 |MOLECULE: CARBOXY-CIS,CIS-MUCONATE CYCLASE; 4: |1l0q-C |30.3 |2.2 |286 |391 | 15 |MOLECULE: SURFACE LAYER PROTEIN; 5: |1l0q-B |30.3 |2.2 |286 |391 | 15 |MOLECULE: SURFACE LAYER PROTEIN; 6: |1l0q-A |30.3 |2.2 |286 |391 | 15 |MOLECULE: SURFACE LAYER PROTEIN; 7: |1l0q-D |30.2 |2.2 |286 |391 | 15 |MOLECULE: SURFACE LAYER PROTEIN; 8: |1qni-D |28.3 |2.8 |317 |572 | 11 |MOLECULE: NITROUS-OXIDE REDUCTASE; 9: |1qni-F |28.0 |2.8 |317 |572 | 11 |MOLECULE: NITROUS-OXIDE REDUCTASE; 10: |1fwx-D |28.0 |2.8 |316 |588 | 10 |MOLECULE: NITROUS OXIDE REDUCTASE; 11: |1fwx-A |27.9 |2.9 |318 |591 |10 |MOLECULE: NITROUS OXIDE REDUCTASE; 21: |1sq9-A |25.0 |3.0 |293 |378 |10 |MOLECULE: ANTIVIRAL PROTEIN SKI8; 23: |2z2o-C |24.5 |2.6 |269 |299 |10 |MOLECULE: VIRGINIAMYCIN B LYASE; 25: |1yfq-A |24.4 |3.1 |287 |342 |11 |MOLECULE: CELL CYCLE ARREST PROTEIN BUB3; 42: |1pev-A |23.5 |2.9 |282 |610 | 9 |MOLECULE: ACTIN INTERACTING PROTEIN 1; 159: |1bpo-B |13.6 |4.1 |261 |493 | 8 |MOLECULE: CLATHRIN; 277: |1a12-B |9.3 |3.9 |235 |401 |8 |MOLECULE: REGULATOR OF CHROMOSOME CONDENSATION 1;
The above table shows some condensed Dali results. The proteins that might have direct carcinogenic effects via the immune system are highlighted. Most of the proteins that have structural similarity to SBP are highly involved in redox reactions,like the reductases.
Back to results Results 2ece
Back to Selenium binding protein