Hypothetical protein Method: Difference between revisions
| Line 1: | Line 1: | ||
=='''Evolution'''== | =='''Evolution'''== | ||
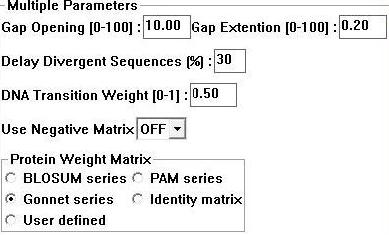
Sequences for evolutionary analysis were gathered from NCBI using | Sequences for evolutionary analysis were gathered from NCBI using BLASTP (refseq). These were selected from different phyla on the basis of sequence identity (>30%). More distantly related sequences were obtained using the Psi-BLAST algorithm available at Interpro. Selected sequences were gathered in FASTA format and input into a .txt file sequentially. ClustalX 1.83 [1] was then used to generate a multiple sequence alignment for the above sequences using the following parameters; | ||
[[Image:clus.jpg]] | [[Image:clus.jpg]] | ||
| Line 16: | Line 16: | ||
A bootstrapped tree representing radiation was then copied to Microsoft power point 2007 to emphasise key features and significant bootstrap values. | A bootstrapped tree representing radiation was then copied to Microsoft power point 2007 to emphasise key features and significant bootstrap values. | ||
=='''Structure'''== | =='''Structure'''== | ||
Revision as of 07:50, 15 June 2009
Evolution
Sequences for evolutionary analysis were gathered from NCBI using BLASTP (refseq). These were selected from different phyla on the basis of sequence identity (>30%). More distantly related sequences were obtained using the Psi-BLAST algorithm available at Interpro. Selected sequences were gathered in FASTA format and input into a .txt file sequentially. ClustalX 1.83 [1] was then used to generate a multiple sequence alignment for the above sequences using the following parameters;
Figure 1: Clustalx 1.83 alignment parameters [1]
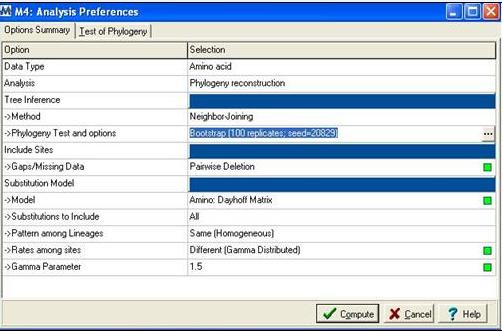
Following alignment; sequences were trimmed in Clustalx [1] from residues 31-184 to remove extending ‘tails’ for more accurate phylogenetic analysis. The alignment file was then saved as an .aln file and exported to MEGA 4.0 [2] using the program’s conversion to MEGA format tool. MEGA [2] was then used to construct a bootstrapped neighbour joining tree using the following parameters;
Figure 2: Mega alignment parameters [2], Gamma parameter was established using Quartet puzzling [3].
A bootstrapped tree representing radiation was then copied to Microsoft power point 2007 to emphasise key features and significant bootstrap values.
Structure
Function
The main methods for the function of our protein involved the us of STRING analysis and Profunc analysis. This involved the submitting of our protein to the respective services, after which they provided their separate anaylsis of the specified protein. Literature was used to confirm any speculations garnished from the results obtained.
Abstract | Introduction | Method |
Results | Discussion | Conclusion | References